Mutations in adenine-binding pockets enhance catalytic properties
advertisement
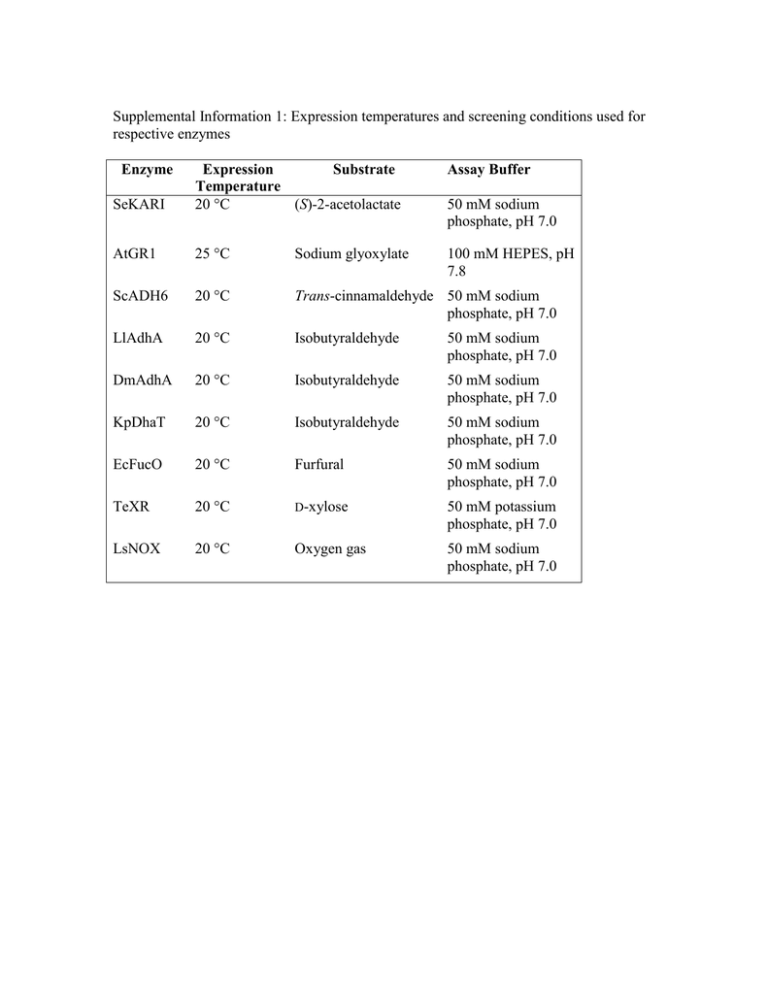
Supplemental Information 1: Expression temperatures and screening conditions used for respective enzymes Enzyme Assay Buffer SeKARI Expression Substrate Temperature 20 °C (S)-2-acetolactate AtGR1 25 °C Sodium glyoxylate 100 mM HEPES, pH 7.8 ScADH6 20 °C Trans-cinnamaldehyde 50 mM sodium phosphate, pH 7.0 LlAdhA 20 °C Isobutyraldehyde 50 mM sodium phosphate, pH 7.0 DmAdhA 20 °C Isobutyraldehyde 50 mM sodium phosphate, pH 7.0 KpDhaT 20 °C Isobutyraldehyde 50 mM sodium phosphate, pH 7.0 EcFucO 20 °C Furfural 50 mM sodium phosphate, pH 7.0 TeXR 20 °C D-xylose 50 mM potassium phosphate, pH 7.0 LsNOX 20 °C Oxygen gas 50 mM sodium phosphate, pH 7.0 50 mM sodium phosphate, pH 7.0 Supplemental Information 2: Brief discussion of the enzymes studied in this work. Saccharomyces cerevisiae cinnamyl alcohol dehydrogenase (ScADH6) is used by yeast for the detoxification of aromatic aldehydes (Larroy et al. 2002). Arabidopsis thaliana glyoxylate reductase (AtGR1) is involved in the glyoxylateglycolate shuttle for the regulation of photosynthesis (G. J. Hoover et al. 2007). Lactococcus lactis and Drosophila melanogaster alcohol dehydrogenases (LlAdhA and DmADH) are promiscuous alcohol dehydrogenases previously investigated for use in microbial isobutanol production (Liu et al. 2012). Klebsiella pneumoniae propanediol dehydrogenase (KpDhaT) is one of the main enzymes for the metabolic pathway in K. pneumoniae that enables the species to metabolize glycerol as a sole source of carbon and energy by reduction of 3hydroxypropanal to propane-1,3-diol (Marcal et al. 2009). Escherichia coli lactaldehyde reductase (EcFucO) catalyzes the inter-conversion between L-lactaldehyde and L-1,2-propanediol during the anaerobic dissimilation of fucose (Cocks et al. 1974) and aerobic growth on L-1,2-propanediol (Chen et al. 1989; Wang et al. 2011). It also reduces furfural, the dehydration product of xylose, an important fermentation inhibitor in sugar syrups derived from woody biomass to the less toxic furfuryl alcohol (Wang et al. 2011). Xylose reductase is the first enzyme of xylose metabolism in fungi; the enzyme from Talaromyces emersonii (TeXR) is a highly active and thermostable member of this family (Fernandes et al. 2009). NADH oxidase from Lactobacillus sanfranciscensis (LsNOX) is an important enzyme in maintaining redox balance (Jansch et al. 2011) and has been used for NAD(P)+ regeneration (Petschacher et al. 2014). relative activity normalized to optimum [%] Supplemental Information 3: pH-activity profiles and thermostability of indicated enzymes. T50 is defined as the temperature at which 50% of the initial activity is retained after 10 min incubation. DmADH WT 100 80 50 mM Citrate buffer 60 50 mM Sodium phosphate buffer 40 50 mM TRIS-HCl buffer 20 50 mM Carbonatebicarbonate buffer 0 2 3 4 5 6 7 8 9 10 11 relative activity normalized to optimum [%] pH DmADH V108I 100 80 50 mM Citrate buffer 60 50 mM Sodium phosphate buffer 40 50 mM TRIS-HCl buffer 20 50 mM Carbonatebicarbonate buffer 0 2 3 4 5 6 7 pH Enzyme DmADH WT T50 (°C) 46.6 ± 0.6 DmADH V108I 48.8 ± 0.8 EcFucO WT 44.7 ± 0.7 EcFucO M185A 45.3 ± 4.0 EcFucO M185C 40.8 ± 2.9 8 9 10 11 Supplemental Information 4: Kinetic parameters of characterized enzymes. Numbers in parentheses refer to α, the Hill constant. Mutation kcat (min-1) NADH NADPH NADH KM (μM) NADPH Substrate AtGR1 Wild Type 7.3 ± 0.9 27 ± 3.9 76 ± 24 20 ± 6 690 ± 160 AtGR1 C68E 2.7 ± 0.2 13 ± 0.8 66 ± 29 10 ± 3 60 ± 11 AtGR1 C68R 13 ± 3.1 7.3 ± 1.8 53 ± 11 24 ± 9 120 ± 46 ScADH6 Wild Type 1300 ± 370 16000 ± 2300 130 ± 51 140 ± 15 170 ± 26 ScADH6 T255K 3200 ± 87 9000 ± 1100 240 ± 13 37 ± 21 56 ± 4 DmADH Wild Type 7.8 ± 1.6 - 57 ± 6 - 130 ± 33 DmADH V108I 9.2 ± 3.4 - - 100 ± 24 EcFucO Wild Type 1.8 ± 0.2 - - 1400 ± 130 EcFucO M185A 6.2 ± 2.0 - - 390 ± 87 EcFucO M185C 6.5 ± 1.8 - - 910 ± 140 LsNOX Wild Type 1200 ± 94 890 ± 170 42 ± 6 70 ± 8 (2.9 ± 0.8) 39 ± 4 (2.7 ± 0.6) 55 ± 2 (3.0 ± 0.3) 85 ± 12 73 ± 60 - LsNOX I122V 4000 ± 240 1400 ± 280 20 ± 9.3 110 ± 64 - LsNOX I155L 2700 ± 520 3500 ± 360 100 ± 80 32 ± 8.4 - LsNOX I243M I122VI155L I22VI243M I155LI243M I122VI155LI243M 2800 ± 370 3100 ± 120 150 ± 175 56 ± 48 - 2700 ± 490 5000 ± 540 70 ± 12 120 ± 30 - 5500 ± 290 4500 ± 49 59 ± 22 31 ± 21 - 2500 ± 290 2400 ± 93 83 ± 7.6 120 ± 24 - 7900 ± 520 11000 ± 360 93 ± 96 140 ± 300 - LsNOX LsNOX LsNOX LsNOX REFERENCES Chen,Y.M., Lu,Z. and Lin,E.C. (1989) J Bacteriol, 171, 6097–6105. Cocks,G.T., Aguilar,T. and Lin,E.C. (1974) J Bacteriol, 118, 83–88. Fernandes,S., Tuohy,M.G. and Murray,P.G. (2009) J Biosci, 34, 881–890. Hoover,G.J., Van Cauwenberghe,O.R., Breitkreuz,K.E., Clark,S.M., Merrill,A.R. and Shelp,B.J.(2007) Can J Botany, 85, 883–895. Jansch,A., Freiding,S., Behr,J. and Vogel,R.F.(2011) Food Microbiol, 28, 29–37. Petschacher,B., Staunig,N., Müller,M., Schürmann,M., Mink,D., De Wildeman,S., Gruber,K. and Glieder,A. (2014) Comput Struct Biotechnol J, 9, e201402005.








