Document
advertisement
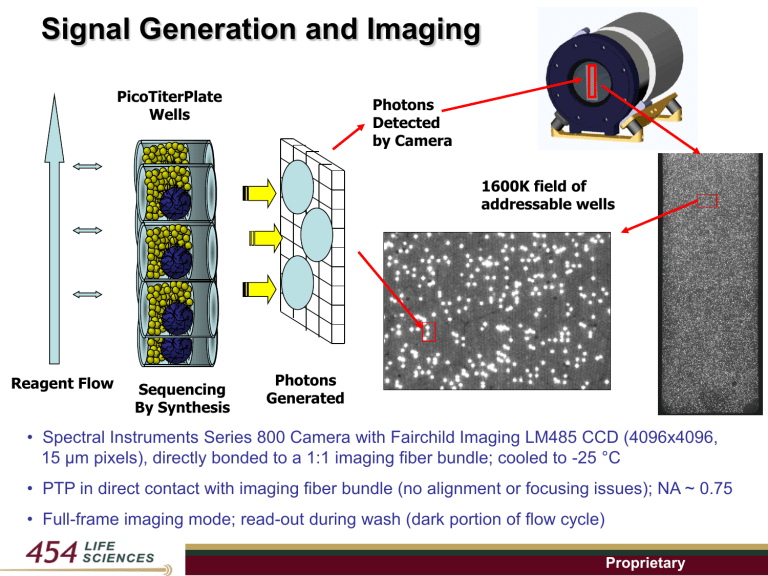
Signal Generation and Imaging PicoTiterPlate Wells Photons Detected by Camera 1600K field of addressable wells Reagent Flow Sequencing By Synthesis Photons Generated • Spectral Instruments Series 800 Camera with Fairchild Imaging LM485 CCD (4096x4096, 15 μm pixels), directly bonded to a 1:1 imaging fiber bundle; cooled to -25 °C • PTP in direct contact with imaging fiber bundle (no alignment or focusing issues); NA ~ 0.75 • Full-frame imaging mode; read-out during wash (dark portion of flow cycle) Proprietary Image Processing Raw data series of images T C G A T Data converted into flowgrams” Proprietary Signal-Processing & Basecalling Image Data → Signal processing Pipeline → Flowgrams → Quality Filtering → HQ Reads → Basecalling → HQ Bases → Mapping & Assembly 4-mer T A Flow C Order G 3-mer TTCTGCGAA 2-mer 1-mer Key sequence = TCAG for well identification and calibration Proprietary Mapping in Flowgram Space Reference Chromosome Flowgram Fragment Flowgrams (RN) …,1, 3, 1, 0, 0, 2, 2, 0, 0, 1, 2, 3, 0, 1, 0… …, 1.00, 3.14, 0.15, 0.20, 0.21, 1.84, 1.95,… Re-sequencing and find variants to the reference genome Proprietary De Novo Assembly in Flowgram Space Overlap to form contigs Fragment Flowgrams (RN) Draft sequences of new genomes (species that have not been sequenced before) Proprietary Potential Sources of Error Optical Cross-Talk • Light penetrates to adjacent fibers; leads to signal contamination Non-DNA Well DNA Well False signal True signal Chemical Cross-Talk • PPi / ATP produced in DNA well appears in down-stream non-DNA wells due to convection / diffusion; leads to signal contamination Flow PPi Non-DNA Well False signal DNA Well True signal Proprietary Signal-Processing Solution Original Filtered (51x51) kernel 2D intensity contour Pixel Pixel Filtering removes signal cross-contamination Proprietary Typical GS20 Run Results 42 Flow Cycles ~ 30MB per run ~ 300,000 reads ~ 100 bp per read # of reads 15000 10000 5000 0 41 51 61 0 1 2 71 81 91 101 111 121 131 60% > 50% wells error-free ~ 1 % individual read error % of reads 50% 40% 30% 20% 10% 0% 3 4 5 6 7 8 9 # of errors per read Proprietary 10+* Consensus Accuracy > 99.99+% when 10x over-sampling Example Ref / assembled genome: 2 -GTGCGCGCGCGGGACTAATCCCGGTTCGCGCGTCGGGCATGACACGCAAC- 10 reads aligned to this position Read #: FlowSignal Read 1: 2.52 Read 2: 1.95 Read 3 2.11 Read 4: 1.53 Read 5: 1.32 Read 6: 2.14 Read 7: 2.06 Read 8: 1.85 Read 9: 2.21 Read 10: 2.17 Consensus (mean): 1.99 Proprietary De Novo Assembly Results 4 runs of GS20 (E. Coli 4,639,675 bp) Each data point represents 1/2 GS20 run 100.00% 1800 # of contigs 1600 1400 # Contigs 90.00% % Coverage 80.00% 1200 70.00% 1000 800 60.00% 600 50.00% 400 200 0 % coverage 2000 40.00% 1 run 4.5 2 runs 9 13 16.5 3 runs 20 23 4 runs 26.5 30.00% 30 Oversampling Proprietary Genome Sequencer FLX Recently Launched Next Generation Sequencing Platform in Q1, 2007 in collaboration with Roche Diagnostics At least 400K (200K) reads of avg. 250 bp (100 bp) 8 hours (5 hours) run time Single read avg accuracy >99.5% (99%) over 200 (100) bases Consensus read accuracy > 99.99% Avg yield from single run ~ 100 Mb (20-30 Mb) (GS20 performance) Improved fluidics for faster reagent delivery Firmware control of reagent delivery & camera timing On-board reagent dilution Optimized biochemistry Improved algorithms with corrections for – Crosstalk (for higher densities) – Signal droop & Phasing Numerical filter for improved rejection of low quality reads Proprietary Fluidics Modification – Air Plug Insertion Camera cover G A C Debubbler Air mM air plug in tube PTP Inlet Nuc Conc (non-dim) PTP Inlet Conc (non-dim) 21 s Injection: Ver 1.0 (New M Concentration Profile Pre-dilution withReagent & w/o air bubble Mode 11 Standard 0.80.8 Air Bubble Insertion 0.60.6 t air bubbles removed at debubbler 0.40.4 0.2 0.2 0 0 0 0 20 20 40 40 Time (s) Time (s) Proprietary 60 60 80 Whole Genome Sequencing Results from GS FLX E. Coli (50% GC) C. jejuni (35%GC) T. thermophilus (71% GC) Proprietary Observed Individual Read Accuracy Cumulative Read Error 4.0% 3.5% E. coli run #1 09_29A E. coli run #2 09_29B 09_14 + 09_18A E. coli run #3 09_18B+09_25 E. coli run #4 Thermophilus T. thermophilus C C.jejuni jejuni Reported in Nature 2005 3.0% 2.5% 2.0% GS20 Q2 2006 1.5% 1.0% 0.5% 0.0% 0 50 100 150 200 250 Base Position All blind-filtered reads (no reference genome required) Proprietary Newbler™ Assembly Results from GS FLX C. jejuni T. thermophilus E. coli E. coli (GS20) Genome Size: 1,641,481 2,127,575 4,639,675 4,639,675 Number of Runs: ½ ½ 1 3 Assembly Contigs: 25 56 105 140 Assembly Cover: 98.31% 98.15% 97.61% 97.46% Overall Accuracy: 99.996% 99.991% 99.998% 99.998% Avg. Contig Size: 64.6 kb 39.8 kb 43.3 kb 32.4 kb N50 Contig Size: 116 kb 82.1 kb 105.5 kb 67.2 kb Largest Contig: 481 kb 383.0 kb 204.7 kb 164 kb Proprietary