document
advertisement

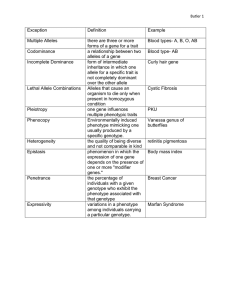
Biology 2250 Principles of Genetics Announcements Lab 4 Information: B2250 (Innes) webpage download and print before lab. Virtual fly: log in and practice http://biologylab.awlonline.com/ Quiz – 3 answers http://webct.mun.ca:8900/ All quizzes on WebCT for Review Office Hours: 1:30 – 2:30 Tue, Wed., Thr or by appointment: 737-4754, dinnes@mun.ca Mendelian Genetics Topics: -Transmission of DNA during cell division Mitosis and Meiosis - Segregation - Sex linkage (problem: how to get a white-eyed female) - Inheritance and probability - Independent Assortment - Mendelian genetics in humans - Linkage - Gene mapping -Gene mapping in other organisms (fungi, bacteria) - Extensions to Mendelian Genetics - Gene mutation - Chromosome mutation (- Quantitative and population genetics) Linkage: Summary • Recombination: generates new combinations (inter and intrachromosomal) • Genetic maps: - genes linked on the same chromosome - location of new genes relative to genes already mapped Linkage: Summary • Hunting for genes (Human Diseases) - genetic markers: DNA variation - co-inheritance with diseases using pedigree information - recombinants used to estimate linkage Extensions to Mendelian Genetics Ch. 14 From Gene to Phenotype Readings: Ch. 14 p. 454 – 473 Problems: Ch. 14: 2, 3, 4, 5, 6, 7 Chapter 1 Genes, environment, organism Phenotype = gene + env. + gene x env. + gene x gene Mendelian Genetics: Genotype Phenotype Dominance ? G x E interaction Extensions to Mendelian Genetics (Gene Phenotype) 1. Dominance 2. Multiple alleles 3. Pleiotropy 4. Epistasis (gene interaction) 5. Penetrance and expressivity Gene interaction 1. Alleles at one gene Dominance 2. Different genes Epistasis 1. Dominance Location of heterozygote between two homozygotes 1. Complete 2. No dominance 3. Incomplete (partial) 4. Codominance Homozygotes: A1A1 A2A2 Heterozygote: A1A2 Incomplete Dominance red white pink Codominance Human Blood Groups: Genotype Phenotype** AA A AB AB co-dominance BB B ** antigen protein on RBC Codominance Molecular Markers Allele A B AB AA BB BB Heterozygote distinguished from homozygotes 2. Multiple Alleles (ABO Blood groups - 3 alleles) Genotype Phenotype (6) (4) --------------------------------------------OO O recessive AA, AO A dominant BB, BO B dominant AB AB co-dominant --------------------------------------------- Multiple alleles in clover Test for Allelism Possibilities: or 1. alleles for the same gene - all crosses show Mendelian ratios (1:1 3:1 1:2:1) 2. more complex inheritance (> 1 gene) Example: white, yellow, pink Cross white x yellow white x pink yellow x pink F1 yellow pink pink 3 alleles: w y p 6 genotypes: w w y y p p F2 3:1 yellow : white 3:1 pink : white 3:1 pink : yellow pw yw yp 3. Pleiotropy (one gene affects > 1 trait) Example: Mouse Gene affects: 1. coat colour ( 2. survival AA Homozygous wildtype dark , yellow) zzz Yellow Parents Crosses A. x -----> all B. x ---> 1/2 1/2 C. x ----> 2/3 1/3 Explanation A. AA B. AA x AA all AA x AYA C. AYA x AYA ½ AYA , ½ AA ¼ AA ½ AYA ¼ AYAY 1 1/3 : 2 2/3 dies Interpretation Gene affects both coat colour and survival 1. AY dominant to A for coat colour 2. AY recessive lethal for survival Pleiotropy Genotype AA A AY AY AY Phenotype coat colour survival dark dark yellow ? alive alive dead G + E = P Trait 1 Pleiotropy Gene A Trait 2 Epistasis Gene A Trait Gene B Gene interaction 4. Epistasis (gene interaction) More than one gene affects a character One gene pair masks or modifies the expression of another gene pair AABB x aabb ----> AaBb x AaBb ---> F2 F1 Dihybrid Epistasis AaBb Gene A and B unlinked x AaBb F2 A- B- 9/16 A- bb 3/16 aa B- 3/16 aa bb 1/16 4 distinct phenotypes (2 traits) (peas: shape, colour) Epistasis: Gene A and Gene B interact phenotype of 1 trait 1. Epistasis (BbEe X BbEe) Labrador retriever Coat Colour (B and E genes) F2 Ratio 9/16 3/16 3/16 1/16 Genotype Phenotype B- Eblack B- ee gold bb Ebrown bb ee gold Gene E allows colour deposition Ratio 9/16 4/16 3/16 Epistasis Allele E Allele B Golden brown B- ee bb ee bb E- black B- E- 2. Epistasis (AaBb X AaBb) Example: Flower petal colour F2 Ratio 9/16 3/16 3/16 1/16 Genotype A- BA- bb aa Baa bb Phenotype Purple White White White Ratio 9/16 7/16 Gene B colourless (white) A-bb aabb Gene A colourless (white) purple aaB- A- B- 5. Penetrance and Expressivity Phenotype: genotype, genetic background, and environment Variable Expression: Penetrance Expressivity Penetrance: percentage of individuals that show some degree of expression of a mutant genotype Example: Polydactyly (P) extra digits pp normal Pp PP 10 % normal polydactyly 90 % polydactyly Expressivity: degree that a given genotype is expressed phenotypically Example: Pp individuals which do express the extra digits can vary (a) extra digit on each hand and foot (b) extra digit on one hand only (c) complete digit or vestige Same genotype Variable expressivity of piebald spotting in beagles Summary - segregation and independent assortment can explain a variety of patterns of genetic variation - Phenotype = Genotype + Environment Genetic interaction: genotype, epistasis, genetic background Mutation Source of genetic variation: Gene Mutation - somatic, germinal Chromosome mutations (Ch. 11 prob. 1, 2) - structure - number Mutation Gene Mutation a+------>a Forward mutation a ------>a+ Reverse mutation 1. Somatic mutation - not transmitted to progeny 2. Germinal Mutation - transmitted to next generation Somatic Mutations Petal colour: Rr red rr white Plant genotype: Rr mutation: Rr rr Somatic mutations Germinal mutations AA (blue) Aa self aa(white) Mutant Phenotypes Morphological Lethal Biochemical Resistance Conditional - DTS (David T. Suzuki) (permissive and restrictive conditions) Mutation Frequency Drosophila eye-colour w+ w 4 x 10-5 per gamete Humans Hemophilia (X-linked recessive) 4 x 10-5 per gamete (1 in 25,000) “It is estimated that up to 30% of cases of hemophilia have no known family history. Many of these cases are the result of new mutations. This means that hemophilia can affect any family.” Mutation Frequency Drosophila eye-colour w+ w 4 x 10-5 per gamete Mutation rate for a particular gene: very low (efficient repair) but, Large number of genes in a genome: mutations occur every generation 4 x 10-5 x 50,000 genes = 2 mutations Gene Mutation Mutations are rare and random Ultimate source of genetic variation Cancer: Proto-oncogene oncogene cancer mutation Chromosome Mutations Gene mutation: detected genetically Chromosome Mutations: detected genetically and cytologically 1. Structure 2. Number Chromosome Mutations 1. Structure Ch. 11 363 – 372 2. Number Ch. 11 p. 350 - 363 1. Chromosome Structure Karyotype: 1. size and number 2. centromere position: telocentric acrocentric metacentric submetacentric acentric (lost) Chromosome Structure 3. Heterochromatin pattern - heterochromatin (dark) - euchromatin (light) 4. Banding patterns: a) staining Giemsa bands b) polytene chromosomes (flies) G-bands Paint of Chr-22 “Paint” Structural Abnormalities Normal 1. Deletion a b c d e f a c d e f 2. Duplication a b b c d e f 3. Inversion c b f a e d 4. Translocation a b c d j k g h i e f Structural Abnormalities 1. Deletions: deletion homozygote---->usually lethal deletion heterozygote----> viable deletion loop (pairing of homologues) b a a c c d d deletion Deletion heterozygote deletion loop Pseudodominance Deletion Heterozygote: deletion loop (pairing of homologues) Phenotype: b a + c + d + deletion + b + + Deletion Mapping Prune pn Structural Abnormalities Deletion: notch-wing (Drosophila) Phenotype Genotype wing survival N+ N+ normal alive N+ N notch alive N N dead (recessive lethal) Genetics of Deletions • Reduced map distance ( chromosome shortened) • Recessive lethal • Deletion loop (detected during meiosis) Structural Abnormalities 2. Duplications: tandem duplication a b b c d maintain original function evolve new function Unequal crossing over deletion Tandem duplication Bar Eye Mutation (Dominant) Gene Duplication and Evolution Gene duplication - Evolution of new function Example: Hemoglobin genes - duplication Express in different stages: embryo – fetus – adult Hemoglobin: Alpha Beta Gamma ……….. Structural Abnormalities 3. Inversions - different gene order - usually viable abcdef abcdef homozygote NN abedcf abedcf abcdef abedcf heterozygote homozygote NI II normal (N) inversion (I) Cytological consequences of an Inversion Heterozygote: Inversion Loop Fig. 11-21 a b c d e a d c b e crossover X Inversion Loop Cytological consequences of an Inversion Heterozygote: Inversion Loop Cross-over within an inversion dicentric bridge (broken) acentric fragment (lost) deletions Inversion heterozygote with crossing over Fig. 11-22 Inversion Heterozygote • Reduced recombination frequency (suppression of crossing over) • Semisterile 4. Translocation a b c d j k g h i e f Translocation Heterozygote (meiosis) N1 N2 T1 T2 Translocation Translocation heterozygote Fig. 11-24 Translocation heterozygote Adjacent segregation T1 N2 N1 T2 inviable Translocation heterozygote Alternate segregation N1 N2 T1 T2 viable Translocation Change linkage relationships (position effects) Change chromosome size Semisterile - unbalanced meiotic products normal Corn Pollen aborted % aborted = ?? Structural Abnormalities Normal 1. Deletion a b c d e f a c d e f 2. Duplication a b b c d e f 3. Inversion c b f a e d 4. Translocation a b c d j k g h i e f