Virus Classification and Description Lec. 3

Virus Classification And Description
Classification Parameters
• Several Parameters Are Used for Classification
– Viral classification study is referred to as Taxonomy
– 73 families exist so far!!
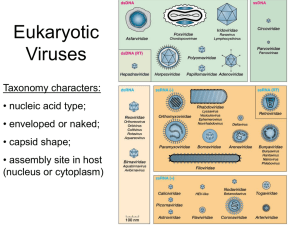
– Type of genomic nucleic acid
– Size of virion and genome
– Capsid structure
– Host
– Replication mechanism
Size of Viruses
• Ranges of sizes
– 20 nm to 500 nm (spherical)
– 12 nm to 300-2000 nm (rod like)
– Easily observed with electron microscope
– Ex.1 Mimivirus is 500 nm
• Infects algae
– Ex.2 Parvovirus is 20 nm in diameter
• Infects algae
– Viral genomes range in size 2,000 bp to 1,200,000 bp
Comparison Between Cellular Genome and Viral Genome
• Viral genome
– 2-1,200 Kb
– Encoded proteins: 2-1,200
– Compact and economical
• Cellular genome
– 3x10 9 bp
– 30,000 proteins
– Massive and with 90% non-coding DNA
ssDNA Viruses
• ssDNA Viruses have the following characteristics
– Small genome, 2-7 Kb
• Possibly due to unstable nature of ssDNA compared to dsDNA
– Circular genomes with the exception of Parvoviridae
(hairpin)
– No envelope
– Predominantly icosahedral capsids
dsDNA Viruses
• Examples of dsDNA viruses that infect humans
– HSV, HPV and adenoviruses
• Among the largest known viruses
• Genome size varies from 5 to 1180 Kb
• Unfragmented genomes
• Both linear and circular
• Large genome size attributed to stability of dsDNA
• Low error rate during replication
• No dsDNA virus is known to infect plants
• Phages are dsDNA viruses (95%)
dsRNA Viruses
• They utilize RNA dependent polymerase
• Icosahedral capsids
• Capsids stays intact inside cell. Why?Genome protection.
• Transcription occurs via viral RNA polymerases
• Reoviruses (dsRNA) are capable of infecting multiple species (plants, vertebrates, fungi). Not a common phenomenon.
• Rhabhoviridae infect multiple species as well
• The fact that they carry their own RNA replication/transcription proteins makes them more adept
Viruses With + strand RNA Genomes
• Very common of plant viruses to be + ssRNA
• Only one phage family is + ssRNA
• RNA viruses have linear genomes
• Similar to ssDNA viruses they are susceptible to nucleases and divalant cation degradation
• Coronavirus has the largest genome of + ssRNA virus (16-30 Kb)
- ssRNA Viruses
• This group includes some of the deadliest viruses
– Ebola, rabies, influenza, measles
• Only helical nucleocapsids
• Nucleocapsid seems to provide stability for RNA dependent RNA polymerase to generate + ssRNA
• + ssRNA=mRNA
Viruses With Reverse Transcription
• 3 families belong to this group
– Retroviridae, Ex. HIV
– Hepadnaviridae, Ex. Hep B
– Caulimoviridae, Ex. Cauliflower Mosaic Virus
• These families utilize enzyme that uses an RNA template to make DNA template
• Reverse transcriptase is packaged in capsid
– Similar to + ssRNA and – ssRNA that package the RNA dependent polymerase
• Retroviruses package 2 copies of their RNA genome in the capsid
Satellite Viruses/Nucleic Acids
• These viruses require a helper virus
• Their genomes encode for capsid proteins
• Nucleic acid satellites are either non-coding or encode for non-capsid proteins
• Mostly a plant phenomenon
• In humans the Hep virus resembles characteristics of satellite virus/viroid
Viroids
• These are plant pathogens
• They replicate in the absence of helper virus
• Genome is circular and single stranded
• 250-400 nt
• Cellular DNA dependent RNA polymerases are used for replication
• High degree of internal base pairing
• RNA behaves as enzyme
– i.e cut/ligate themselves
– Ribozymes