copies/ml (log10) - National Institute for Biological Standards and
advertisement

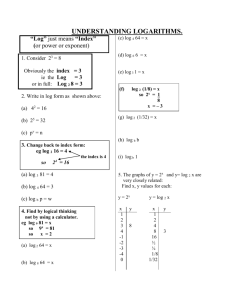
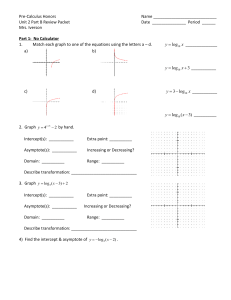
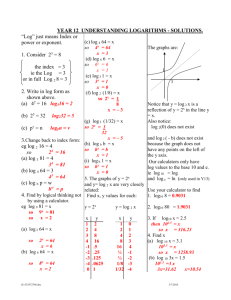
Potential uses of EBV and CMV viral load assays In solid organ and hematopoietic stem cell transplantation As triggers for pre-emptive therapy for disease prevention For disease diagnosis For treatment monitoring As surrogate markers of anti-viral resistance For safety monitoring in clinical trials (new immunosuppressive agents) Other Disease diagnosis and treatment monitoring other EBVrelated disease- nasopharyngeal carcinoma ,NK and Hodgkin’s lymphoma Population based screening- congenital CMV disease CMV and EBV Viral Load Assays Current Problems Many “In-house” ; not standardized or cross referenced Optimal sampling site uncertain - serum, Leukocytes/lymphocytes, whole blood Optimal sampling frequency uncertain Natural history studies are scarce so that “trigger points” for intervention have not been clearly defined Development of an International Standard for EBV and CMV Viral Load Assessment Dr Jutta K Preiksaitis Provincial Public Health Laboratory (Alberta) University of Alberta Edmonton and Calgary, Alberta Canada On behalf of the American Society of Transplantation Infectious Diseases Community of Practice and the Canadian Society of Transplantation Objective of Study To examine the inter-laboratory variability in qualitative and quantitative CMV and EBV viral load assessments Funded by the American Society of Transplantation and the Canadian Society of Transplantation ( armslength educational grant Roche Canada) Coordinated through the American Society of Transplantation Infectious Diseases Community of Practice CMV Viral Load Assays Establishing the “expected result” Viral stock (purified nucleocapsids of Merlin, a clinical isolate in human in CMV seronegative human plasma) Quantified by nucleocapsid count using electron microscopy log 10 copies/ml =4.52 Calculation of a mean of replicate viral load results from seven reference laboratories (included use of all available commercial assays) log 10 copies/ml =5.0 Panel Design 12 samples 2 negatives (CMV seronegative plasma) 7 samples -dilutions of purified viral stock; replicates of two dilutions were included 3 clinical samples (1:30 dilution in CMV seronegative plasma) UL54 mutation (not ganciclovir resistant) UL97mutation (ganciclovir resistant) and gB mutation No mutation CMV PCR Methods Utilized n=35 panels (33 labs) 19 US, 12 Canada , 2 EU Other commercially available kits (ASRs) 23% Roche Amplicor 26% Inhouse 51% Results Summary 35 panels / 33 laboratories CMV DNA Copies/ml (log10) 7.00 6.50 6.00 5.50 5.00 4.50 4.00 3.50 3.00 2.50 2.00 1.50 1.00 0.50 0.00 02 09 Clinical sample 07 08 04 11 03 12 06 CMV Sample Number CMV viral panel sample Expected result based on stock quantified by reference laboratories Positive but not quantifiable (assigned lowest detectable value) 10 05 01 Summary of CMV Qualitative Results (constructed samples) 35 panels / 33 labs Sample No EM –based expected result copies/ml (log10) Reference lab expected result copies/ml (log10) 02 0.0 †09 Number of panels Negative (%) Positive-NQ (%) Positive-Q (%) 0.0 34 (97) 0 1 (3) 0.0 0.0 33 (94) 0 1 (3) 07 1.5 2.0 26 (74) 6 (17) 3 (9) 08 2.5 3.0 4 (11) 4 (11) 27 (77) 04 3.5 4.0 0 1 (3) 34 (97) 11 3.5 4.0 0 2 (6) 33 (94) 03 4.5 5.0 0 0 35 (100) 12 4.5 5.0 0 0 35 (100) 06 5.5 6.0 0 0 35 (100) † One test was invalid Pos-NQ: positive but not quantifiable Pos-Q: positive with quantifiable results Summary of CMV Quantitative results (constructed samples) 35 panels / 33 laboratories Sample No EM –based expected result copies/ml (log10) Reference lab expected result copies/ml (log10) 07 1.5 2.0 08 2.5 3.0 04 3.5 4.0 11 3.5 4.0 03 4.5 5.0 12 4.5 06 5.5 Number positive †GM SD copies/ml (log10) Median (range) copies/ml (log10) 9 2.2 0.44 0 (0-2.78) 31 3.1 0.58 3.01 (0-4.32) 35 3.89 0.52 4.02 (2.33-5.08) 35 3.84 0.52 3.95 (2.62-5.01) 35 4.83 0.44 4.89 (3.42-5.89) 5.0 35 4.80 0.49 4.90 (3.68-5.91) 6.0 35 5.59 0.52 5.51 (4.65-6.73) † Geometric mean; negative results were excluded CMV quantitative results relative to expected result [reference labs as “gold” standard] §Number of panel results falling within specified parameter relative to expected result [reference labs] (copies/ml, log10) Sample No # positive log±0.2 (%) log±0.5 (%) log±1 (%) > log±1 (%) 07 9 2 (22) 7 (78) 9 (100) 0 08 31 8 (26) 21 (68) 27 (87) 4 (13) 04 35 17 (49) 26 (74) 33 (94) 2 (6) 11 35 16 (46) 25 (71) 32 (91) 3 (9) 03 35 19 (54) 25 (71) 34 (97) 1 (3) 12 35 16 (46) 25 (71) 32 (91) 2 (6) 06 35 7 (20) 15 (43) 32 (91) 3 (9) §negative results were excluded CMV Qualitative and Quantitative results (clinical samples) 35 panels / 33 laboratories Clinical Sample Number Qualitative Result Negative (%) Pos-NQ (%) Pos-Q (%) Quantitative Result copies/ml (log10) †GMSD Median (range) #10 #05 #01 13 (37) 0 0 9 (26) 1 (3) 0 13 (37) 34 (97) 35 (100) 2.78 0.72 3.89 0.53 3.97 0.47 2.24 (0-4.18) 3.87 (2.73-4.89) 3.99 (3.08-5.05) † GM=Geometric mean; negative results were excluded Result linearity over dynamic range (each line represents results from one lab) CMV copies/ml (log10) using PCR Commercial assays (Lab =17) In-house assays (Lab =18) 7.00 7.00 6.00 6.00 5.00 5.00 4.00 4.00 3.00 3.00 2.00 2.00 1.00 1.00 0.00 0 0.00 0 1 2 3 4 5 6 7 1 2 CMV Copies/ml (log10) Expected result 3 4 5 6 7 Comparison of Intra and Inter laboratory variation in CMV vial load assessments in duplicate specimens mean coefficient of variation (CV), % Duplicate samples (sample 04 and 11) 35 panels Duplicate samples (sample 03 and 12) 35 panels p value* Intra-Lab 21.48 17.62 0.720 Inter-Lab 149.23 139.15 0.316 p value* < 0.0001 < 0.0001 * Fisher Exact Test (two tailed) CMV Conclusions Significant variation exists in reported results. The greatest variation was observed in clinical samples and constructed samples with low viral load. As viral load increased, there was less variation independent of the assay platforms used (commercial versus inhouse) False negative results were not observed in samples with viral load greater than 3.0 log copies/ml (expected result) even when lower limit of detection reported was higher than this value Variation is lower limits of detection may have significant impact on duration of treatment based on recommendation of treatment until viral load is non-detectable If ± 0.5 log10 is considered “acceptable” assay variation, only 62.5 % of the results observed fell within this range CMV Conclusions As a group, commercial assays demonstrated overall less variability compared to all “in house” developed assays, but some of the former have limitations related to lower sensitivity and limited dynamic range Inter-laboratory variability was significantly greater than intra-laboratory variability, highlighting the need for an international reference standard for assay calibration EBV Viral Load Assays Establishing the “expected result” EBV viral stock (Namalwa cell line in EBV seronegative plasma) Quantified by Namalwa cell count using assumption of 2 EBV genome copies per cell Calculation of a geometric mean of replicate viral load results from seven reference laboratories ( included use of all available commercial assays) Panel Design 12 samples Constructed samples-(total cell count in each sample fixed to mimic total white cell count in normal peripheral blood) 2 negatives ( EBV-negative Molt-3 cells in EBV seronegative plasma) 7 samples -dilutions of EBV-positive Namalwa cells and EBV-negative Molt-3 cells ; two dilutions were replicated 3 clinical plasma samples (diluted in EBV seronegative plasma) Two patients had EBV-positive B cell post-transplant lymphoproliferative disorder EBV PCR Methods Utilized n=30 panels (28 labs) 16 US, 11 Canada, 2 EU Other commercially available kits (ASRs) 40% Inhouse 60% Results Summary 30 panels / 28 laboratories 7.00 EBV DNA copies/ml (log10) 6.00 5.00 4.00 3.00 2.00 1.00 0.00 01 08 09 03 05 Clinical Samples EBV Viral Panel Samples 10 02 11 06 EBV Sample Number Gold Standard Expected Result Based on Cell Count Positive but not quantifiable (assigned lowest detectable value) 07 04 12 Summary of EBV Qualitative Results (constructed samples) 30 panels reported from 28 laboratories Sample No. Expected result based on Namalwa cell count copies/ml (log10) Number of panels Negative (%) Positive-NQ (%) Positive-Q (%) 01 0.0 30 (100) 0 0 08 0.0 28 (93) 0 2 (7) 09 1.3 27 (90) 1 (3) 2 (7) 03 2.3 16 (53) 3 (10) 11 (37) 05 3.3 3 (10) 2 (7) 25 (83) 10 3.3 3 (10) 1 (3) 26 (87) 02 4.3 0 1 (3) 29 (97) 11 4.3 0 2 (7) 28 (93) 06 5.3 0 1 (3) 29 (97) Quantitation based on cell count Pos-NQ: positive, not quantifiable Pos-Q: positive, quantifiable Summary of EBV Quantitative results (Constructed Samples) 30 panels reported from 28 labs Sample No. 09 Expected result based on Namalwa cell count copies/ml (log10) 1.3 Number of positive results † GM SD copies/ml (log10) Median (range) copies/ml (log10) 3 1.890.93 0.00 (0.002.74) 14 2.480.59 0.00 (0.003.78) 03 2.3 05 3.3 27 2.970.52 2.92 (0.00-4.14) 10 3.3 27 3.020.61 2.92 (0.00-4.12) 02 4.3 30 3.920.59 4.03 (2.76-5.04) 11 4.3 30 3.880.66 06 5.3 30 4.810.81 †Geometric mean; negative results were excluded 3.97 (2.18-5.00) 4.96 (2.15-6.09) EBV quantitative results (constructed samples) relative to expected result [Namalwa cell count as “gold” standard] §Number of panel results falling within specified parameter relative to expected result [Namalwa cell count] (copies/ml, log10) Sample No Number positive results log±0.2 (%) log±0.5 (%) log±1 (%) > log±1 (%) 09 3 0 1 (33) 1 (33) 2 (67) 03 14 3 (21) 10 (71) 12 (86) 2 (14) 05 27 5 (19) 16 (59) 26 (96) 1 (4) 10 27 6 (22) 14 (52) 25 (93) 2 (7) 02 30 10 (33) 17 (63) 25 (83) 5 (17) 11 30 8 (27) 15 (50) 25 (83) 5 (17) 06 30 4 (13) 17 (57) 25 (83) 5 (17) § negative results were excluded EBV Qualitative and Quantitative results (clinical samples) 30 panels reported from 28 labs Clinical Sample Number Qualitative Result #07 #04 #12 Negative (%) 0 0 0 Pos-NQ (%) 0 0 0 30 (100) 30 (100) 30 (100) †GMSD 4.08 0.60 3.95 0.56 4.21 0.61 Median (range) 4.09 (3.09- 5.12) 3.96 (3.10 – 5.31) 4.36 (3.08 – 5.12) Pos-Q (%) Quantitative Result copies/ml, log10 † GM= Geometric mean Result linearity over dynamic range (each line represents results from one lab) EBV copies/ml (log10) using PCR Commercial assays (Lab =12) In-house assays (Lab = 18) 7.00 7.00 6.00 6.00 5.00 5.00 4.00 4.00 3.00 3.00 2.00 2.00 1.00 1.00 0.00 0.00 1.00 2.00 3.00 4.00 5.00 0.00 6.00 0.00 1.00 2.00 3.00 4.00 EBV copies/ml expected quantification based on cell count 5.00 6.00 Comparison of Intra and Inter laboratory variation in EBV vial load assessments in duplicate specimens Mean coefficient of variation (CV), % Duplicate (sample 05 and 10) 25 panels Duplicate (sample 02 and 11) 30 panels p value* Intra-Lab 39.01 30.48 0.234 Inter-Lab 135.56 135.26 1.0 p value* < 0.0001 < 0.0001 * Fisher Exact Test (two tailed) Conclusions Significant variation in reported results exists in all samples independent of viral load and of assay platforms used (commercial versus in-house) If ± 0.5 log10 is considered “acceptable” variation in a Q NAT assay, our results indicate that only 56 % of all results fell within that parameter Greater QNAT variations were observed in cellular constructed samples when compared to acellular plasma samples indicating that DNA extraction in cellular samples may need further improvement Inter-laboratory variability was significantly greater than intra-laboratory variability, highlighting the need for an international reference standard for assay calibration Next Steps Highest Priority Establishment of an international reference standard for EBV and CMV qualitative and quantitative assay calibration Acknowledgments Technical Committee Dr Dr Dr Dr Xiao-Li Pang Julie Fox Geraldine Miller Angie Caliendo Technical and other support Jayne Fenton Sandra Shokopoles Kim Martin Ana Shynader AST ID Community of Practice Dr John Saldanha Dr Alan Heath Participating Laboratories Canada Children’s Hospital of British Columbia, Vancouver St. Paul’s Hospital, Vancouver Provincial Laboratory for Public Health Alberta, Edmonton & Calgary National Microbiology Laboratory, Winnipeg St. Joseph’s Health Care, Hamilton Hospital for Sick Children, Toronto Mt. Sinai Hospital, Toronto Children’s Hospital of Eastern Ontario, Ottawa London Laboratory Services, London St. Justine Hospital, Montreal Centre hospitalier de l'Université Laval, Quebec City QE II Health Sciences Centre, Halifax Newfoundland Public Health Laboratory, St. John’s Europe Erasmus MC, University Medical Center Rotterdam, The Netherlands Institute for Medical Microbiology, Basel, Switzerland USA UCLA Healthcare Clinical Labs, Los Angeles Stanford Hospital and Clinics, Stanford Yale-New Haven Hospital, New Haven Emory Hospital, Atlanta University of Iowa, Iowa City University of Chicago Hospitals, Chicago Johns Hopkins Hospital, Baltimore University of Michigan Medical Center, Ann Arbor Warde Medical Laboratory, Ann Arbor Mayo Clinic, Rochester St. Louis Children’s Hospital, St. Louis Viracor Laboratories, Lee’s Summit University of North Carolina Hospital, Chapel Hill Mt. Sinai Hospital, New York Cleveland Clinic, Cleveland Oregon Health & Science University, Portland Children’s Hospital of Pittsburgh, Pittsburgh Vanderbilt University Medical Center, Nashville Seattle Cancer Care Alliance, University of Washington, Seattle Children’s Hospital, Birmingham