DNA replication
advertisement

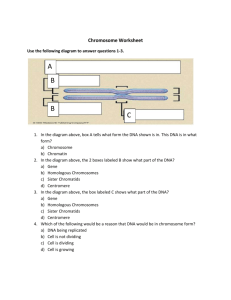
DNA, Chromosomes and DNA Replication Dr.Aida Fadhel Biawi DNA REPLICATION DNA replication is a biological process that occurs in all living organisms and copies their DNA ;it is the basis for biological inheritance . The process starts when one double-stranded DNA molecule produces two identical copies of the molecule . How does DNA replicate? DNA Replication is a semiconservative process that results in a doublestranded molecule that synthesizes to produce two new double stranded molecules such that each original single strand is paired with one newly made single strand . Semiconservative replication would produce two copies that each contained one of the original strands and one new strand . "replication begins at specific sites on DNA molecule called "origins of replication, origins are specific sequence of bases mammalian DNA have many origins The replication fork is a structure that forms within the nucleus during DNA replication. It is created by helicases, which break the hydrogen bonds holding the two DNA strands together. Replication fork is where the parental DNA strands hasn't untwist. Replication bubbles allow DNA replication to speed up therefore the untwisted DNA would not be attacked by enzymes while replicating .. ( Which enzymes can attack DNA?? ) Specific enzymes & proteins recognize origins & bind DNA : 1- primase and DNA polymerase will find these specific portions and will bind to the template DNA at the correct location . ( DNA replication requires a RNA primer , primer synthesized by the enzyme primase , primer is a short strand RNA about 5 bases and RNA primer is complementary to DNA ) - new DNA synthesized by DNA polymerase , DNA polymerase binds to parent DNA strand with primer. - DNA polymerase sequentially adds deoxyribonucleotides to RNA primer , deoxyribonucleotides added have bases complementary to parent strand DNA . - The rate nucleotide additions in bacteria add about 500 bases/second while in mammels add about 50 bases/second ??!! 2-replication requires strand separation a. strand separation begins at origin of replication (Helicase) b. specific proteins prevent the two separated DNA strands from coming back together (single strand binding protein) At origin of replication ,one strand of DNA is made in a continuous manner (the leading strand) and the other in a discontinuous manner (the lagging strand(. DNA is made in only the 5-prime to 3-prime direction and the replication bubble opens the original double stranded DNA to expose both a 3-prime to 5-prime template (Leading strand template) and it complement . The lagging strand must be synthesized as a series of discontinuous segments of DNA .?? These small fragments are called Okazaki fragments and they are joined together by an enzyme known as DNA ligase . Enzyme Function in DNA replication DNA Helicase Also known as helix destabilizing enzyme. Unwinds the DNA double helix at the Replication Fork. DNA Polymerase Builds a new duplex DNA strand by adding nucleotides in the 5' to 3' direction. Also performs proof-reading and error correction. Single-Strand Binding (SSB) Proteins Bind to ssDNA and prevent the DNA double helix from re-annealing after DNA helicase unwinds it thus maintaining the strand separation. Topoisomeras e Relaxes the DNA from its super-coiled nature. DNA Ligase Primase Re-anneals the semi-conservative strands and joins Okazaki Fragments of the lagging strand. Provides a starting point of RNA (or DNA) for DNA polymerase to begin synthesis of the new DNA strand. DNA CONDENSATION A cell's genetic information, in the form of DNA, is stored in the nucleus. The space inside the nucleus is limited and has to contain billions of nucleotides that compose the cell's DNA. Therefore, the DNA has to be highly organized or condensed. There are several levels to the DNA packaging . At the finest level, the nucleotides are organized in the form of linear strands of double helices. The DNA strand is wrapped around histones, a form of DNA binding proteins. Each unit of DNA wrapped around a histone molecule is called a nucleosome . The nucleosomes are linked together by the long strand of DNA . Characteristics of Histone Proteins - Octamere structure - Responsible for packaging DNA into nucleosomes - 4 different types: H2A, H2B, H3, H4 Nucleosome Structures Histone octamer 2 H2A 2 H2B 2 H3 2 H4 Beads on a String—10 nm Fiber protein purification histones (= 1g per g DNA) 09/01/12 H1 •Basic (arg, lys); •+ charges bind H3 to - phosphates H2A on DNA H2B H4 SBL201 DNA 22 To further condense the DNA material, nucleosomes are compacted together to form chromatin fibers .The chromatin fibers then fold together into large looped domain .During the mitotic cycle, the looped domains are organized into distinct structures called the chromosomes . Packing of DNA into Chromatin - histone-DNA complexes, referred to as nucleosomes, which are further folded into higher-order chromatin structures . Nucleofilament or solenoid 30 nm fiber Fig. 9 Orders of chromatin structure from naked DNA to chromatin to fully condensed chromosomes... - Chromosomes are also used as a way of referring to the genetic basis of an organism as either diploid or haploid .Many eukaryotic cells have two sets of the chromosomes and are called diploid. Other cells that only contain one set of the chromosomes are called haploid . • A diploid cell has two sets of each of its chromosomes • A human has 46 chromosomes (2n = 46) • In a cell in which DNA synthesis has occurred all the chromosomes are duplicated and thus each consists of two identical sister chromatids Maternal set of chromosomes (n = 3) 2n = 6 Paternal set of chromosomes (n = 3) Two sister chromatids of one replicated chromosome Centromere Two nonsister chromatids in a homologous pair Pair of homologous chromosomes (one from each set) Chromosome Duplication • In preparation for cell division, DNA is replicated and the chromosomes condense • Each duplicated chromosome has two sister chromatids, which separate during cell division An eukaryotic cell has multiple chromosomes, one of which is represented here. Before duplication, each chromosome has a single DNA molecule. Once duplicated, a chromosome consists of two sister chromatids connected at the centromere. Each chromatid contains a copy of the DNA molecule. Mechanical processes separate the sister chromatids into two chromosomes and distribute them to two daughter cells. 0.5 µm Chromosome duplication (including DNA synthesis) Centromere Separation of sister chromatids Centrometers Sister chromatids Sister chromatids Chromosome Duplication • Because of duplication, each condensed chromosome consists of 2 identical chromatids joined by a centromere. • Each duplicated chromosome contains 2 identical DNA molecules (unless a mutation occurred), one in each chromatid: Non-sister chromatids Centromere Duplication Sister chromatids Two unduplicated chromosomes Sister chromatids Two duplicated chromosomes Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Structure of Chromosomes • The centromere is a constricted region of the chromosome containing a specific DNA sequence, to which is bound 2 discs of protein called kinetochores. • Kinetochores serve as points of attachment for microtubules that move the chromosomes during cell division.???!!! Metaphase chromosome Centromere region of chromosome Kinetochore Kinetochore microtubules Sister Chromatids Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Chromosome structure p: short arm q: long arm C: constriction point or centromere , the location of centromere give the chromosome its shape and can be used to help describe the location of the genes. A typical mitotic chromosome at metaphase – Diploid - A cell possessing two copies of each chromosome (human body cells). • Homologous chromosomes are made up of sister chromatids joined at the centromere. – Haploid - A cell possessing a single copy of each chromosome (human sex cells). Phases of the Cell Cycle • Interphase – G1 - primary growth – S - genome replicated – G2 - secondary growth • M - mitosis • C - cytokinesis Cell cycle begins with the formation of two cells from the division of a parent cell and ends when the daughter cell does so as well. Observable under the microscope, M phase consists of two events, mitosis (division of the nucleus) and cytokinesis (division of the cytoplasm). As replication of the DNA occurs during S-phase, when condensation of the chromatin occurs two copies of each chromosome remain attached at the centromere to form sister chromatids. After the nuclear envelope fragments, the microtubules of the mitotic spindle separate the sister chromatids and move them to opposite ends of the cell. Cytokinesis and reformation of the nuclear membranes occur to complete the cell division. -Most of the time, cells are in interphase, where growth occurs and cellular components are made. DNA is manufactured during S phase. -To prepare the cell for S phase (DNA synthesis), G1 phase occurs (the preparation of DNA synthesis machinery, production of histones). -In an analogous manner, the cell prepares for mitosis in the G2 phase by producing the machinery required for cell division. -The length of time spent in G1 is variable. In growing mammalian cells often spend ??? hours in G1 phase. G2 is usually shorter than G1 and is usually ??? hours. And S phase ??? Interphase • G1 - Cells undergo majority of growth • S - Each chromosome replicates (Synthesizes) to produce sister chromatids – Attached at centromere – Contains attachment site (kinetochore) • G2 - Chromosomes condense - Assemble machinery for division such as centrioles Mitosis Some haploid & diploid cells divide by mitosis. Each new cell receives one copy of every chromosome that was present in the original cell. Produces 2 new cells that are both genetically identical to the original cell. DNA duplication during interphase Mitosis Diploid Cell