T cell antigen receptors PPT
advertisement

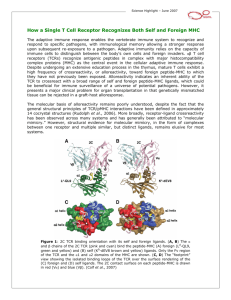
Topic 6 The T Cell Antigen Receptor Complex ©Dr. Colin R.A. Hewitt crah1@le.ac.uk What you should know by the end of this lecture • Each clone of T cells expresses a single TcR specificity • How the TcR was discovered • The similarities and differences between TcR and antibodies • The structure and organisation of the TcR genes • Somatic recombination in TcR genes • Generation of diversity in TcR • Structure function relationship of TcR • Why TcR do not undergo somatic mutation Discovery of the T cell antigen receptor (TcR) Polyclonal T cells from an immunised strain A mouse Grow and clone a single antigenspecific T cell in-vitro with antigen, IL-2 and antigen presenting cells Monoclonal (cloned) T cells In vitro “clonal selection” means each daughter cell has the same antigen specificity as the parent cell Most molecules present on the monoclonal T cells will be identical to the polyclonal T cells EXCEPT for the antigen combining site of the T cell antigen receptor Making anti- clonotypic TcR antibodies T cell clone from a strain A mouse Naïve strain A mouse Make monoclonal antibodies by hybridisation of the spleen cells with a myeloma cell line The strain A mouse will not make antibodies to the hundreds of different molecules associated with strain A T cells due to self tolerance BUT The naïve mouse has never raised T cells with the specificity of the T cell clone, SO the only antigen in the immunisation that the A strain mouse has never seen will be the antigen receptor of the monoclonal T cells Making anti- clonotypic TcR antibodies Screen the supernatant of each cloned hybridoma against a panel of T cell clones of different specificity (i.e.cells with subtly different antigen-binding structures) Monoclonal antibodies YYY Clone used for immunisation T cell clones Y Y Y Anti-TcR Abs that recognise only one clone of T cells are CLONOTYPIC Hypothesise that anti-clonotype Abs recognise the antigen receptor Discovery of the T cell antigen receptor (TcR) Y Y YYYYYYY Lyse cells and add anti-clonotype Ab that binds to unique T cell structures YYYY Capture anti-clonotype Ab-Ag complex on insoluble support IMMUNOPRECIPITATION Wash away unbound protein YYYY Elute Ag from Ab and analyse the clonotypically-expresssed proteins biochemically Principal component was a heterodimeric 90kDa protein composed of a 40kDa and a 50kDa molecule ( and chains) Several other molecules were co-immunoprecipitated. Structure of the TcR polypeptides T cell clone A T cell clone B T cell clone C Intact TcR chain polypeptides Cyanogen bromide digestion of the and proteins Biochemical analysis of digestion products C V C V C V Polypeptides contain a variable, clone-dependent pattern of digestion fragments and a fragment common to all TcR Cloning of the TcR genes T B The experimental strategy • The majority of genes expressed by T and B lymphocytes will be similar • Genes that greatly differ in their expression are most likely to be directly related to the specialised function of each cell • Subtract the genes expressed by B cells from the genes expressed by T cells leaving only the genes directly related to T cell function Cloning of TcR genes by subtractive hybridisation T B mRNA AAAAA AAAAA T cell single stranded cDNA Hybridise the AAAAA cDNA and mRNA shared between T and B cells Digest unhybridised B cell mRNA AAAAA Discard hybrids AAAAA AAAAA Isolate non-hybridising material specific to T cells Clone and sequence T cellspecific genes Analysis of T cell-specific genes Of the T cell-specific genes cloned, which cDNA encoded the TcR? Assumptions made after the analysis of Ig genes: TcR genes rearrange from germline configuration Ig gene probes can be used as TcR genes will be homologous to Ig genes Restriction enzyme sites 32P V D J C GERMLINE DNA C REARRANGED DNA 32P V DJ Find two restriction sites that flank the TcR region Cut the T cell cDNA and placental (i.e. germline) DNA and Southern blot the fragments The TcR genes rearrange, but are not immunoglobulin genes Gel electrophoresis followed by Southern blot using a TcR probe Placenta Size of digested genomic DNA B T Rearranged allele The T cell antigen receptor Antigen combining site Resembles an Ig Fab fragment VL VH V L VH CL CH CH CH CH V V CL Fab Fc CH CH Domain structure: Ig gene superfamily Monovalent Carbohydrates No alternative constant regions C C Hinge + + Cytoplasmic tail + Transmembrane region Never secreted Heterodimeric, chains are disuphidebonded Very short intracytoplasmic tail Positively charged amino acids in the TM region Antigen combining site made of juxtaposed V and V regions 30,000 identical specificity TcR per cell CH CL VH VLof Ig C C V V of the TcR View structures T cell antigen receptor diversity • Unlike MHC molecules TcR are highly variable in the individual • Diversity focused on small changes in the charge & shape presented at the end of the T cell receptor. • TcR diversity to the peptide antigens that bind to MHC molecules • Mechanisms of diversity closely related to T cell development • Random aspects of TcR construction ensures maximum diversity • Mechanisms of diversity generation similar to immunoglobulin genes Generation of diversity in the TcR COMBINATORIAL DIVERSITY Multiple germline segments In the human TcR Variable (V) segments: ~70, 52 Diversity (D) segments: 0, 2 Joining (J) segments: 61, 13 The need to pair and chains to form a binding site doubles the potential for diversity JUNCTIONAL DIVERSITY Addition of non-template encoded (N) and palindromic (P) nucleotides at imprecise joints made between V-D-J elements SOMATIC MUTATION IS NOT USED TO GENERATE DIVERSITY IN TcR Organisation of TcR genes L&V x70-80 C J x 61 TcR L&V x52 D1 J1 x 6 C1 D2 J2 x 7 TcR TcR genes segmented into V, (D), J & C elements (VARIABLE, DIVERSITY, JOINING & CONSTANT) Closely resemble Ig genes (~IgL and ~IgH) This example shows the mouse TcR locus C2 TcR gene rearrangement by SOMATIC RECOMBINATION Vn V2 V1 J Germline TcR Rearranged TcR 1° transcript Spliced TcR mRNA Rearrangement very similar to the IgL chains C TcR gene rearrangement RESCUE PATHWAY There is only a 1:3 chance of the join between the V and J region being in frame Vn+1 Vn V2 V1 J C chain tries for a second time to make a productive join using new V and J elements Productively rearranged TcR 1° transcript TcR gene rearrangement SOMATIC RECOMBINATION L & V x52 D1 J C1 D2 J C2 Germline TcR D-J Joining V-DJ joining Rearranged TcR 1° transcript C-VDJ joining Spliced TcR mRNA TcR gene rearrangement RESCUE PATHWAY There is a 1:3 chance of productive D-J rearrangement and a 1:3 chance of productive D-J rearrangement (i.e only a 1:9 chance of a productive chain rearrangement) V D1 J C1 D2 J C2 Germline TcR D-J Joining V-DJ joining 2nd chance at V-DJ joining Need to remove non productive rearrangement Use (DJC)2 elements V, D, J flanking sequences Sequencing upstream and downstream of V, D and J elements revealed conserved sequences of 7, 23, 9 and 12 nucleotides. V 7 23 9 V 7 23 9 12 9 7 9 D 7 12 9 12 7 J 9 23 7 J Recombination signal sequences (RSS) HEPTAMER - Always contiguous with coding sequence 9 V 7 23 √ V 9 7 12 23 7 D 7 9 9 12 9 9 12 7 D NONAMER - Separated from the heptamer by a 12 or 23 nucleotide spacer 7 J 23 7 12 9 √ 9 23 7 J 12-23 RULE – A gene segment flanked by a 23mer RSS can only be linked to a segment flanked by a 12mer RSS Molecular explanation of the 12-23 rule 12-mer = one turn 23-mer = two turns 23 V 7 Intervening DNA of any length 9 12 9 7 DJ Molecular explanation of the 12-23 rule V4 V1 V8 V9 V3 V2 V7 V6 V3 V4 V2 V5 9 9 23-mer • Heptamers and nonamers align back-to-back 7 7 V7 V8 V9 12-mer V1 V6 Loop of intervening DNA is excised DJ • The shape generated by the RSS’s acts as a target for recombinases V5 DJ • An appropriate shape can not be formed if two 23-mer flanked elements attempted to join (i.e. the 12-23 rule) Junctional diversity Mini-circle of DNA is permanently lost from the genome 9 7 V 7 12 23 9 9 23 Coding joint 7 7 12 9 Signal joint DJ VDJ Imprecise and random events that occur when the DNA breaks and rejoins allows new nucleotides to be inserted or lost from the sequence at and around the coding joint. Non-deletional recombination V1 V1 V1 V3 V2 7 V2 V3 9 23 V4 23 V4 7 V9 DJ 9 V9 V4 Looping out works if all V genes are in the same transcriptional orientation 9 DJ 12 7 D J How does recombination occur when a V gene is in opposite orientation to the DJ region? 9 12 7 DJ Non-deletional recombination 9 23 7 V4 9 12 7 D J 1. 2. 9 7 V4 23 9 3. V4 and DJ in opposite transcriptional orientations 23 9 23 7 V4 7 V4 4. 9 23 7 V4 9 12 7 D J 2. 1. 9 9 23 7 12 9 23 9 V4 12 7 7 V4 D J Heptamer ligation - signal joint formation 7 D J 3. V4 9 23 9 4. 9 12 23 7 7 D J 7 7 12 9 V to DJ ligation coding joint formation V4 D J Fully recombined VDJ regions in same transcriptional orientation No DNA is deleted Steps of TcR gene recombination V 7 23 V 7 23 D J 9 12 7 7 23 9 12 7 9 7 23 D J The two RAG1/RAG 2 complexes bind to each other and bring the V region adjacent to the DJ region 9 9 12 7 9 12 7 V 9 Recombination activating gene products, (RAG1 & RAG 2) and ‘high mobility group proteins’ bind to the RSS 9 • The recombinase complex makes single stranded nicks in the DNA, the ends of each broken strand. • The nicks are ‘sealed’ to form a hairpin structure at the end of the V and D regions and a flush double strand break at the ends of the heptamers. • The recombinase complex remains associated with the break D J Steps of TcR gene recombination V 7 23 9 D J 9 12 7 V D J D J 9 12 7 7 23 9 V A number of other proteins, (Ku70:Ku80, XRCC4 and DNA dependent protein kinases) bind to the hairpins and the heptamer ends. The hairpins at the end of the V and D regions are opened, and exonucleases and transferases remove or add random nucleotides to the gap between the V and D region DNA ligase IV joins the ends of the V and D region to form the coding joint and the two heptamers to form the signal joint. Junctional diversity: P nucleotide additions 7 23 V AT GTGACAC J D TA CACTGTG 9 9 12 7 V TC CACAGTG AG GTGTCAC 7 7 9 23 12 9 The recombinase complex makes single stranded nicks at random sites close to the ends of the V and D region DNA. TC AG TC CACAGTG AG GTGTCAC 7 GTGACAC CACTGTG 7 V V AT AT J JDTA DTA 9 23 12 9 The 2nd strand is cleaved and hairpins form between the complimentary bases at ends of the V and D region. D J V3 V2 V4 CACAGTG GTGTCAC 7 GTGACAC CACTGTG 7 9 23 12 V5 9 V9 Heptamers are ligated by DNA ligase IV TC AG V AT J DTA V V8 V7 TC AG V and D regions juxtaposed AT TA V6 D J Generation of the palindromic sequence V V V TC AG TC AG TC~GA AG AT TA D J AT TA D J Regions to be joined are juxtaposed Endonuclease cleaves single strand at random sites in V and D segment The nicked strand ‘flips’ out AT TA~TA D J The nucleotides that flip out, become part of the complementary DNA strand In terms of G to C and T to A pairing, the ‘new’ nucleotides are palindromic. The nucleotides GA and TA were not in the genomic sequence and introduce diversity of sequence at the V to D join. Junctional Diversity – N nucleotide additions V TC~GA CACTCCTTA AT AG TTCTTGCAA TA~TA D J Terminal deoxynucleotidyl transferase (TdT) adds nucleotides randomly to the P nucleotide ends of the singlestranded V and D segment DNA V TC~GA CACACCTTA AT AG TTCTTGCAA TA~TA D J Complementary bases anneal V TC~GACACACCTTA D J Exonucleases nibble back free ends V TC CACACCTTA TC~GA GTT ATAT AT AGC TTCTTGCAA TA TA~TA AG D J DNA polymerases fill in the gaps with complementary nucleotides and DNA ligase IV joins the strands TTCTTGCAA TA~TA Junctional Diversity V TCGACGTTATAT AGCTGCAATATA D J TTTTT Germline-encoded nucleotides TTTTT Palindromic (P) nucleotides - not in the germline TTTTT Non-template (N) encoded nucleotides - not in the germline Creates an essentially random sequence between the V region, D region and J region in beta chains and the V region and J region in alpha chains. How does somatic recombination work? 1. How is an infinite diversity of specificity generated from finite amounts of DNA? Combinatorial diversity and junctional diversity 2. How do V region find J regions and why don’t they join to C regions? 12-23 rule 3. How does the DNA break and rejoin? Imprecisely, with the random removal and addition of nucleotides to generate sequence diversity. Why do V regions not join to J or C regions? V D J C IF the elements of the TcR did not assemble in the correct order, diversity of specificity would be severely compromised 2x DIVERSITY Full potential of the beta chain for diversity needs V-D-J-C joining - in the correct order 1x DIVERSITY Were V-J joins allowed in the beta chain, diversity would be reduced due to loss of the imprecise join between the V and D regions Location of junctional diversity TcR chain TcR chain CDR3 CDR 1 CDR2 V-D Join Variability V-J Join Amino acid No. of TcR chain CDR = Complemantarity determining region D-J join Location of junctional diversity in TcR 2 1 3 2 3 1 CDR’s TcRV monomer TcR chain The trimolecular complex MHC class I and TcR V/V MHC class II TcR / V and V of TcR recognising a peptide from MHC class I ribbon plot TcR recognising a peptide from MHC class II ribbon plot Turn through 90º V and V of TcR recognising a peptide from MHC class I wire plot showing amino acid sidechains TcR recognising a peptide from MHC class II wire plot showing amino acid sidechains TcR contact and anchor residue side chains interact with side chains of TcR Hypervariable loops - CDRs 1/2 /3 1/2 1/2 /3 1/2 The most variable loops of the TcR - the CDR3 interact with the most variable part of the MHC-peptide complex CDR’s 1 and 2 interact largely with the MHC molecule View structures T cell co-receptor molecules Lck PTK TcR 3 CD8 Lck PTK TcR CD4 2 MHC Class I MHC Class II CD4 and CD8 can increase the sensitivity of T cells to peptide antigen MHC complexes by ~100 fold CD8 and CD4 contact points on MHC class I and class II CD8 binding site MHC class I CD8 binding site MHC class II TcR TcR-CD3 complex CD3 CD3 The intracytoplasmic region of the TcR chain is too short to transduce a signal The CD3 or (zeta) chains are required for cell surface expression of the TcR-CD3 complex and signalling through the TcR Signalling is initiated by aggregation of TcR by MHC-peptide complexes on APC Transduction of signals by the TcR CD3 ITAMs The cytoplasmic domains of the CD3 complex contain 10 Immunoreceptor Tyrosine -based Activation Motifs (ITAMS) - 2 tyrosine residues separated by 9-12 amino acids - YXX[L/V]X6-9YXX[L/V] As with B cell receptors, immunoreceptor tyrosine-based activation motifs (ITAMs) are involved in the transmission of the signals from the receptor and require clustering of TcR/CD3 and the CD4 or CD8 co-receptors Phosphorylation by Src kinases Kinase domain Enzyme domain that phosphorylates tyrosine residues (to give phosphotyrosine) SH2 domain SH3 domain Unique region Phosphotyrosine receptor domain Adaptor protein recruitment domain ITAM binding domain • Phosphorylation changes the properties of a protein, by changing its conformation • Changes in conformation can activate or inhibit a biochemical activity, or create a binding site for other proteins • Phosphorylation is rapid and requires no protein synthesis or degradation to change the biochemical activity of a target protein • It is reversible via the action of phosphatases that remove phosphate Regulation of Src kinases Kinase domain Inhibitory tyrosine residue SH2 domain SH3 domain Unique region Activating tyrosine residue Phosphorylation of ‘Activating Tyrosine’ stimulates kinase activity Kinase domain SH2 domain SH3 domain Unique region Phosphorylation of ‘Inhibitory Tyrosine’ inhibits kinase activity by blocking access to the Activating Tyrosine Residue Early T cell activation MHC II MHC II CD4 CD45 Receptor associated kinases accumulate under the membrane in close proximity to the cytoplasmic domains of the TcR CD3 complex As the T cell antigen receptor binds the MHC-peptide antigen, the phosphatase CD45 activates kinases such as Fyn P Fyn Lck Zap-70 This mechanism of activation is similar to the used to activate Syk in B cells CD4 CD45 MHC II T cell activation Fyn phosphorylates the ITAMs of CD3, , and ITAMS The tyrosine kinase ZAP-70 binds to the phosphorylated ITAMs of CD3 - further activation requires ligation of the co-receptor, CD4 P Fyn Lck Zap-70 T cell activation Binding of CD4 co-receptor to MHC class II brings Lck into the complex, which then phosphorylates and activates ZAP-70 MHC II ZAP-70 phosphorylates LAT and SLP-76 P SLP-76 P Fyn P LAT P P Lck Zap-70 Activated ZAP-70 phosphorylates LAT & SLP-76 Tyrosine rich cell membrane associated Linker of Activation in T cells (LAT) and SLP-76 associate with cholesterol-rich lipid rafts T cell activation MHC II Activated ZAP-70 phosphorylates Guanine-nucleotide exchange factors (GEFS) that in turn activate the small GTP binding protein Ras Ras activates the MAP kinase cascade P SLP-76 P Fyn Lck P Tec Tec LAT P P SLP76 binds Tec kinases and activates phospholipase C- (PLC-) Zap-70 PLC- cleaves phosphotidylinositol bisphosphate (PIP2) to yield diacylglycerol (DAG) and inositol trisphosphate (IP3) Transmission of signals from the cell surface to the nucleus Almost identical to transmission in B cells • T cell-specific parts of the signalling cascade are associated with receptors unique to T cells - TcR, CD3 etc. • Subsequent signals that transmit signals to the nucleus are common to many different types of cell. • The ultimate goal is to activate the transcription of genes, the products of which mediate host defence, proliferation, differentiation etc. Once the T cell-specific parts of the cascade are complete, signalling to the nucleus continues via three common signalling pathways via: 1.The mitogen-activated protein kinase (MAP kinase) pathway 2.An increase in intracellular calcium ion concentration mediated by IP3 3.The activation of Protein Kinase C mediated by DAG Simplified scheme linking antigen recognition with transcription of T cell-specific genes • MAP Kinase cascade Small G-protein-activated MAP kinases found in all multicellular animals activation of MAP kinases ultimately leads to phosphorylation of transcription factors from the AP-1 family such as Fos and Jun. • Increases in intracellular calcium via IP3 IP3, produced by PLC-, binds to calcium channels in the ER and releases intracellular stores of Ca++ into the cytosol. Increased intracellular [Ca++] activate a phospatase, calcineurin, which in turn activates the transcription factor NFAT. • Activation of Protein Kinase C family members via DAG DAG stays associated with the membrane and recruits protein kinase C family members. The PKC, serine/threonine protein kinases, ultimately activate the transcription factor NFkB The activated transcription factors AP-1, NFAT and NFkB induce B cell proliferation, differentiation and effector mechanisms Estimate of the number of human TcR and Ig Excluding somatic hypermutation Element Immunoglobulin H k TcR Variable segments 40 59 52 ~70 Diversity segments 27 2 D segments in all 3 frames Yes 0 - Yes 0 - Joining segments 6 2 9 (1)* 13 2 Joints with N & P nucleotides No. of V gene pairs 2360 Junctional diversity ~1013 ~1016** Total diversity 61 1 3640 ~1013 ~1016 * Only half of human k chains have N & P regions **No of distinct receptors increased further by somatic hypermutation Why do TcR not undergo somatic mutation? Antigen presentation Foreign antigen APC Y T cell help Y B T Self Antigen Anergy or deletion of anti-self cells Y T No T cell help Y B Antibody Affinity maturation due to somatic mutation Why do TcR not undergo somatic mutation? Y Y B B Y B Y B Y B Affinity maturation due to somatic mutation Y Y B B Occasional B cell that somatically mutates to become self reactive The lack of somatic mutation in TcR helps to prevent autoimmunity T cell help Y No T cell help X B Occasional B cell that somatically mutates to become self reactive Y T Y T T cell that doesn’t mutate T cell that mutates can can not help the may help the self reactive self reactive B cell B cell Autoantibody production If TcR did undergo somatic mutation: TcR interacts with entire top surface of MHC-peptide antigen complex Somatic mutation in the TcR could mutate amino acids that interact with the MHC molecule causing a complete loss of peptide-MHC recognition If TcR did undergo somatic mutation: TcR-MHC interaction is one of many between the T cell and APC On-off rate of TcR determines rate of ‘firing’ to give qualitatively different outcomes Must be of relatively low affinity as cells with high affinity TcR are deleted to prevent self reactivity. If TcR underwent affinity maturation, they would be deleted Why do B cell receptors need to mutate? Neutralisation of bacterial toxins Ab-Ag interaction must be of high affinity to capture and neutralise toxins in extracellular fluids ` Toxin binding blocked Prevents toxicity ` Y There is a powerful selective advantage to B cells that can somatically mutate their receptors to increase affinity SOMATIC MUTATION An alternative TcR: Discovered as Ig-homologous, rearranging genes in non TcR T cells V V V V V J C Human locus 3x D 3x J 1x C 12x V1 3x J C1 2x J C2 The locus is located between the V and J regions V to J rearrangement deletes D, J and C TcR cells can not express TcR Few V regions, but considerable junctional diversity as chain can use 2 D regions T cells Distinct lineage of cells with unknown functions 1-5% of peripheral blood T cells In the gut and epidermis of mice, most T cells express TcR Ligands of TcR are unknown Possibly recognise: Antigens without involvement of MHC antigens - CD1 Class IB genes Summary • The TcR was discovered using clonotypic antibodies • Antibodies and TcR share many similarities, but there are significant differences in structure and function • The structure and organisation of the TcR genes is similar to the Ig genes • Somatic recombination in TcR genes is similar to that in Ig genes • The molecular mechanisms that account for the diversity of TcR include combinatorial and junctional diversity • TcR do not somatically mutate • The highly variable CDR loops map to the distal end of the TcR • The most variable part of the TcR interacts with the peptide