ODU presentation
advertisement

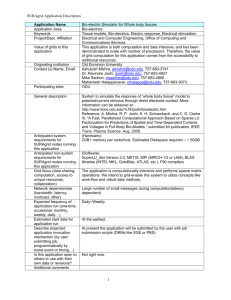
Applications
BioSim
Mahantesh Halappanavar,
Ashutosh Mishra, Ravindra Joshi,
Mike Sachon
SURAgrid “All Hands” Meeting, Washington DC
March 14 – 16, 2007
1
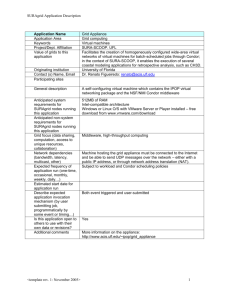
BioSim: Bio-electric Simulator
for Whole Body Tissues
Numerical simulations for electrostimulation of tissues and
whole-body biomodels
Predicts spatial and time dependent currents and voltages
in part or whole-body biomodels
Numerous diagnostic and therapeutic applications, e.g.,
neurogenesis, cancer treatment, etc.
Fast parallelized computational approach
2
Simulation Models
Whole-body discretized within a cubic space
simulation volume
From electrical standpoint, tissues are characterized
as conductivities and permittivities
Cartesian grid of points along the three axes. Thus, at
most a total of six nearest neighbors
* Dimensions in millimeters
3
Numerical Models
Kirchhoff’s node analysis
[( A / L)d{V } / dt {V }( A / L)] 0
Recast to compute matrix only once
[ M ][V |t dt V |t ] [ B(t )]
For large models, matrix inversion is
intractable
LU decomposition of the matrix
4
Numerical Models
Voltage: User-specified timedependent waveform
Impose boundary conditions
locally
Actual data for conductivity
and permittivity
Results in extremely sparse
(asymmetric) matrix
[M]
Red: Total elements in the matrix
Blue: Nonzero Values
5
Why Focus on Solvers?
Scaling: (Source: David Keys, NIA Nov 2006)
–
–
–
–
“Science” phase scales as: O(N )
3
2
O
(
N
)
“Solver” phase scales as
Computation will be almost all solver after several doublings
Optimal solver O(N ) saves computation cycles for physics
6
The Landscape of Sparse Ax=b Solvers
Direct
A = LU
Nonsymmetric
Symmetric
positive
definite
More Robust
Iterative
y’ = Ay
More General
Pivoting
LU
GMRES,
QMR, …
Cholesky
Conjugate
gradient
More Robust
Less Storage
Source: John Gilbert, Sparse Matrix Days in MIT 18.337
7
LU Decomposition
Source: Florin Dobrian
8
LU Decomposition
Source: Florin Dobrian
9
Computational Complexity
100 X 100 X 10 nodes: ~75 GB of memory (8-B floating
precision)
Sparse data structure: ~ 6 MB (in our case)
Sparse direct solver: SuperLU-DIST
– Xiaoye S. Li and James W. Dimmel, “SuperLU-DIST: A Scalable
Distributed-Memory Sparse Direct Solver for Unsymmetric Linear
Systems”, ACM Trans. Mathematical Software, June 2003, Volume
29, Number 2, Pages 110-140.
Fill reducing orderings with Metis
– G. Karypis and V. Kumar, “A fast and high quality multilevel
scheme for partitioning irregular graphs”, SIAM Journal on
Scientific Computing, 1999, Volume 20, Number 1.
10
Performance on compute clusters
144,000-node Rat Model
Blue: Average iteration time
Cyan: Factorization time
11
Output: Visualization with MATLAB
Potential Profile at a depth of 12mm
12
Output: Visualization with MATLAB
Simulated Potential Evolution
Along the Entire 51-mm Width of the Rat Model
13
Deployment on
Mileva: 4-node cluster dedicated for SURAgrid
purposes
Authentication
– ODU Root CA
– Cross certification with SURA Bridge
– Compatibility of accounts for ODU users
Authorization
Initial Goals:
– Develop larger whole-body models with greater resolution
– Scalability tests
14
Grid Workflow
Establish user accounts for ODU users
– SURAgrid Central User Authentication and
Authorization System
– Off-line/Customized (e.g., USC, LSU)
Manually launch jobs based on remote resource
– SSH/GSISSH/SURAgrid Portal
– PBS/LSF/SGE
Transfer files
– SCP/GSISCP/SURAgrid Portal
15
Recent Efforts in grid-enabling:
Porting to 100% open source tools
(GCC/GFORTRAN)
SURAgrid Sites:
– Texas A&M University: Calclab
– University of Virginia: Grid04 and Grid11
Experiments with MUMPS 4
– Symmetric matrices and out-of-core
Acknowledgements:
– Jim Jokl, Steve Losen, Steve Johnson, Brain Brooks,
Kate Barzee and Mary Fran Yafchak
16
News:
(February 14, 2007)
17
18
Conclusions
Science:
– Electrostimulation has variety of diagnostic and
therapeutic applications
– While numerical simulations provide many advantages
over real experiments, they can be very arduous
Grid enabling:
– New possibilities with grid computing
– Grid-enabling an application is complex and time
consuming
– Security is nontrivial
19
Future Steps
Grid-enable BioSim
–
–
–
–
–
Explore alternatives for grid enabling BioSim
Explore funding opportunities
Load Balancing
Establish new collaborations
Scalability experiments with large compute clusters
accessible via SURAgrid
Future applications:
– Molecular and Cellular Dynamics
– Computational Nano-Electronics
– Tools: Gromacs, DL-POLY, NAMD
20
References and Contacts
A Mishra, R Joshi, K Schoenbach and C Clark, “A
Fast Parallelized Computational Approach Based on
Sparse LU Factorization for Predictions of Spatial
and Time-Dependent Currents and Voltages in FullBody Biomodels”, IEEE Trans. Plasma Science,
August 2006, Volume 34, Number 4.
http://www.lions.odu.edu/~rjoshi/
Ravindra Joshi, Ashutosh Mishra, Mike Sachon,
Mahantesh Halappanavar
– (rjoshi, amishra, msachon, mhalappa)@odu.edu
21
Teaching Initiative
CS775/875: Distributed Computing
Ravi Mukkamala
Professor, Department of Computer Science
22
Details:
Graduate course with ~15 students
Guest lecture
Followed by a homework
– Familiarize with grid computing concepts
– Hands-on approach
– Experiment with Globus services & commands
Acknowledgements:
– Jim Jokl, Steve Losen, Steve Johnson, Brain
Brooks, Nicole Geiger, Kate Barzee and Mary
Fran Yafchak
23
24
Conclusions:
Laboratory for testing the concepts
Potential to attract students
For SURAgrid
–
–
–
–
Large number of short-lived certificates
Cleanup … (CRLs?/home drives/…)
Centralized account creation (Still painful )
Short term funding/internships for grad/under-grad
students?
25
THANK YOU !!!
26