20061206-SURAgrid-SGER-YAchak
advertisement

Application Driven Design for
a Large-Scale, Multi-Purpose
Grid Infrastructure
Mary Fran Yafchak, maryfran@sura.org
SURA IT Program Coordinator, SURAgrid project manager
About SURA
501(c)3 consortium of research universities
Major programs in:
– Nuclear Physics (“JLab”, www.jlab.org)
– Coastal Science (SCOOP, scoop.sura.org)
– Information Technology
• Network infrastructure
• SURAgrid
• Education & Outreach
SURA Mission:
foster excellence in scientific research
strengthen the scientific and technical
capabilities of the nation and of the Southeast
provide outstanding training opportunities for
the next generation of scientists and engineers
http://www.sura.org
About SURA
501(c)3 consortium of research universities
Major programs in:
– Nuclear Physics (“JLab”, www.jlab.org)
– Coastal Science (SCOOP, scoop.sura.org)
– Information Technology
• Network infrastructure
• SURAgrid
• Education & Outreach
SURA Mission:
foster excellence in scientific research
strengthen the scientific and technical
capabilities of the nation and of the Southeast
provide outstanding training opportunities for
the next generation of scientists and engineers
http://www.sura.org
Scope of the SURA region
62 diverse member institutions
Geographically - 16 states plus DC
Perspective extends beyond the membership
– Broader education community
• Non-SURA higher ed, Minority Serving Institutions, K-12
– Economic Development
• Regional network development
• Technology transfer
• Collaboration with Southern Governors’ Association
About SURAgrid
A open initiative in support of regional strategy and
infrastructure development
– Applications of regional impact are key drivers
Designed to support a wide variety of applications
– “Big science” but “smaller science” O.K. too!
– Applications beyond those typically expected on grids
• Instructional use, student exposure
• Open to what new user communities will bring
– On-ramp to national HPC & CI facilities (e.g., Teragrid)
Not as easy as building a community or projectspecific grid but needs to be done…
About SURAgrid
Broad view of grid infrastructure
Facilitate seamless sharing of resources within a
campus, across related campuses and between
different institutions
– Integrate with other enterprise-wide middleware
– Integrate heterogeneous platforms and resources
– Explore grid-to-grid integration
Support range of user groups with varying
application needs and levels of grid expertise
– Participants include IT developers & support staff,
computer scientists, domain scientists
SURAgrid Goals
To develop scalable infrastructure that leverages
local institutional identity and authorization while
managing access to shared resources
To promote the use of this infrastructure for the
broad research and education community
To provide a forum for participants to share
experience with grid technology, and participate in
collaborative project development
SURAgrid Vision
SURAgrid
Institutional
Resources
(e.g. Current participants)
Gateways to National
Cyberinfrastructure
(e.g. Teragrid)
VO or Project
Resources
(e.g, SCOOP)
Industry Partner
Coop Resources
(e.g. IBM partnership)
Other externally
funded resources
(e.g. group proposals)
Heterogeneous Environment to Meet Diverse User Needs
Project-Specific View
“MySURAgrid” View
Project-specific tools
SURAgrid
Resources
SURAgrid Resources
and Applications
Sample User Portals
SURA regional development:
• Develop & manage partnership relations
• Facilitate collaborative project development
• Orchestrate centralized services & support
• Foster and catalyze application development
• Develop training & education (user, admin)
• Other…(Community-driven, over time…)
Bowie State
GMU
SURAgrid Participants
UMich
UMD
UKY
(As of November 2006)
UVA
GPN
UArk
Vanderbilt
ODU
UAH
USC
NCState
MCSR
SC
TTU
Clemson
TACC
LATech
= SURA Member
UFL
UAB
TAMU
Kennesaw
State
LSU
ULL
Tulane
= Resources on the grid
GSU
UNCC
Major Areas of Activity
Grid-Building (gridportal.sura.org)
– Themes: heterogeneity, flexibility, interoperability
Access Management
– Themes: local autonomy, scalability, leveraging enterprise
infrastructure
Application Discovery & Deployment
– Themes: broadly useful, inclusive beyond typical users and
uses,promoting collaborative work
Outreach & Community
– Themes: sharing experience, incubator for new ideas,
fostering scientific & corporate partnerships
Major Areas of Activity
Grid-Building (gridportal.sura.org)
– Themes: heterogeneity, flexibility, interoperability
Access Management
– Themes: local autonomy, scalability, leveraging enterprise
infrastructure
Application Discovery & Deployment
– Themes: broadly useful, inclusive beyond typical users and
uses,promoting collaborative work
Outreach & Community
– Themes: sharing experience, incubator for new ideas,
fostering scientific & corporate partnerships
SURAgrid Application Strategy
Provide immediate benefit to applications while
applications drive infrastructure development
Leverage initial application set to illustrate benefits
and refine deployment
Increase quantity and diversity of both applications
and users
Develop processes for scalable, efficient deployment;
assist in “grid-enabling” applications
Efforts significantly bolstered through NSF award:
“Creating a Catalyst Application Set for the Development of
Large-Scale Multi-purpose Grid Infrastructure” (NSF-OCI-054555)
Creating a Catalyst Application Set
Discovery
Ongoing methods: meetings, conferences, word of mouth
Formal survey of SURA members to supplement methods
Evaluation
Develop criteria to help prioritize and direct deployment efforts
Determine readiness to deploy and tools/assistance required
Implementation
Exercise and evolve existing deployment & support processes in
response to lessons learned
Document and disseminate lessons learned
Explore means to assist in grid-enabling applications
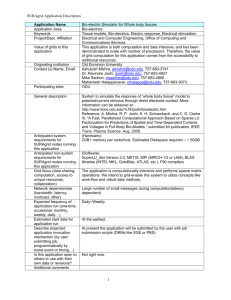
Some Application Close-ups
In SURAgrid demo area today:
GSU: Multiple Genome Alignment on the Grid
– Demo’d by Victor Bolet, Art Vandenberg
UAB: Dynamic BLAST
– Demo’d by: Enis Afgan, John-Paul Robinson
ODU: Bioelectric Simulator for Whole Body Tissues
– Demo’d by Mahantesh Halappanavar
NCState: Simulation-Optimization for Threat
Management in Urban Water Systems
– Demo’d by Sarat Sreepathi
UNC: Storm Surge Modeling with ADCIRC
– Demo’d by Howard Lander
GSU Multiple Genome Alignment
Sequence Alignment Problem
Used to determine biological meaningful relationship among
organisms
– Evolutionary information
– Diseases, causes and cures
– Information about a new protein
Especially compute intensive for long sequences
–
–
–
–
Needleman and Wunsch (1970) - optimal global alignment
Smith and Waterman (1981) - optimal local alignment
Taylor (1987) - multiple sequence alignment by pairwise alignment
BLAST trades off optimal results for faster computation
Examples of Genome Alignment
Alignment 1
Sequence X
Sequence Y
Score
A T A – A G T
A T G C A G T
1 1 -1 -2 1 1 1
Total Score = 2
Alignment 2
Sequence X
Sequence Y
Score
A T A A G T
A T G C A G T
1 1 -1 -1 -1 -1 -1
Total Score = -3
Based on pairwise algorithm
–
–
SM reduced by storing only non-zero elements
–
–
–
Similarity Matrix, SM, built to compare all sequence positions
Observation that many “alignment scores” are zero value
Row-column information stored along with value
Block of memory dynamically allocated as non-zero element found
Data structure used to access allocated blocks
Parallelism introduced to reduce computation
Ahmed, N, Pan, Y, Vandenberg, A and Sun, Y, "Parallel Algorithm for Multiple Genome Alignment
on the Grid Environment," 6th Intl Workshop on Parallel and Distributed Scientific and
Engineering Computing (PDSEC-05) in conjunction with (IPDPS-2005) April 4-8, 2005.
Similarity Matrix Generation
Align Sequence X: TGATGGAGGT
Sequence Y: GATAGG
1 = matching; 0 = non-matching
ss = substitution score; gp = gap score
Generate SM max score with respect to neighbors:
Trace sequences
Back trace matrix to
find sequence matches
Parallel distribution of multiple
sequences
Seq 1-2
Sequences 1-6
Sequences 7-12
Seq 3-4
Seq 5-6
Convergence - collaboration
Algorithm implementation
– Nova Ahmed, Masters CS (now PhD student GT)
– Dr. Yi Pan, Chair Computer Science
NMI Integration Testbed program
– Georgia State, Art Vandenberg, Victor Bolet, Chao
“Bill” Xie, Dharam Damani, et al.
– University of Alabama at Birmingham, John-Paul
Robinson, Pravin Joshi, Jill Gemmill,
SURAgrid
– Looking for applications to demonstrate value
Algorithm Validation: Shared Memory
500
Computation
Time
SGI Origin 2000
24 250MHz R10000; 4G
Limitations
Memory
(Max sequence is
2000 x 2000)
Processors
(Policy limits student
to 12 processors)
Not scalable
Computation Time
(Shared Memory)
400
300
200
100
0
2
4
6
8
10
12
Computation Time
(Shared Memory)
Number of Processors
Performance Validates Algorithm
Computation time decreases with
increased number of processors
Shared Memory vs. Cluster, Grid Cluster*
UAB cluster: 8 node Beowulf (550MHz Pentium III; 512 MB RAM)
Clusters retain algorithm improvement
400
Genome length 3000(Grid)
300
Genome length 3000(Cluster)
200
Genome length 3000( Shared Memory)
100
0
Number of Processors
26
20
14
8
2
Computation
Time (seconds)
500
* NB: Comparing clusters with shared memory is, of
course, relative; systems are distinctly different.
600
400
Genome length 10000 (Grid)
200
Genome length 10000( Cluster)
0
26
Nu mber o f Processo rs
20
14
8
2
Computation
Time (seconds)
Grid (Globus, MPICH-G2) overhead negligible
Advantages of grid-enabled cluster:
Scalable – Can add new cluster nodes to the grid
Easier job submission – Don’t need account on every node
Scheduling is easier – Can submit multiple jobs at one time
500
Speed up (1 cpu / N cpu)
9
Single Cluster
8
400
7
Single Clustered
Grid
300
Speed up
Computation time (sec)
Computation Time
Multi Clustered
Grid
200
100
6
Single Cluster
5
4
3
Single Clustered
Grid
2
Multi Clustered Grid
1
0
0
5
10
15
20
25
30
0
0
Number of processors
5
10
15
20
25
Number of Processors
9 processors available in Multi Clustered Grid
32 processors for other configs.
Interesting: When multiple clusters used (application spanned three
separate clusters), performance improved additionally?!
30
Grid tools used
Globus Toolkit - built on the Open Grid Services
Architecture (OGSA)
Nexus - Communication library, allows multi-method
communication with a single API for a wide range of
protocols. Using Nexus, Message Passing Interface
MPICH-G2 used in the experiments.
Resource specification language (RSL) - job submission
and execution (globus-job-submit, globus-job-run) and
status (globus-job-status)
The Grid-enabling Story
Iterative, evolutionary, collaborative
– 1st ssh to resource and get code working
– 2nd submit from local account to remote globus machine
– 3rd run from SURAgrid portal
SURAgrid infrastructure components providing
improved work-flow
Integration with campus components enables more
seamless access
Overall structure can be used as model for campus
research infrastructure:
– Integrated authentication/authorization
– Portals for applications
– Grid administration/configuration support
SURAgrid Portal
QuickTime™ and a
TIFF (Uncompressed) decompressor
are needed to see this picture.
SURAgrid MyProxy service
QuickTime™ and a
TIFF (Uncompressed) decompressor
are needed to see this picture.
Get Proxy
MyProxy… secure grid credential
QuickTime™ and a
TIFF (Uncompressed) decompressor
are needed to see this picture.
GSI proxy credentials are loaded into your account…
SURAgrid Portal file transfer
QuickTime™ and a
TIFF (Uncompressed) decompressor
are needed to see this picture.
Job submission via Portal
QuickTime™ and a
TIFF (Uncompressed) decompressor
are needed to see this picture.
QuickTime™ and a
TIFF (Uncompressed) decompressor
are needed to see this picture.
Output retrieved
QuickTime™ and a
TIFF (Uncompressed) decompressor
are needed to see this picture.
QuickTime™ and a
TIFF (Uncompressed) decompressor
are needed to see this picture.
SURAgrid Account Management
list my
users
QuickTime™ and a
TIFF (LZW) decompressor
are needed to see this picture.
QuickTime™ and a
TIFF (LZW) decompressor
are needed to see this picture.
SURAgrid Account Management
add user
QuickTime™ and a
TIFF (LZW) decompressor
are needed to see this picture.
Multiple Genome Alignment &
SURAgrid
Collaborative cooperation
Convergence of opportunity
Application / Infrastructure drivers interact
Emergent applications:
–
–
–
–
–
Cosmic ray simulation (Dr. Xiaochun He)
Classification/clustering (Dr. Vijay Vaishnavi, Art Vandenberg)
Muon detector grid (Dr. Xiaochun He)
Neuron (Dr. Paul Katz, Dr. Robert Calin-Jageman, Chao “Bil”l Xie)
AnimatLab (Dr. Don Edwards, Dr. Ying Zhu, David Cofer, James Reid)
IBM System p5 575 with Power5+ Processors
BioSim: Bio-electric Simulator
for Whole Body Tissues
Numerical simulations for electrostimulation of tissues and
whole-body biomodels
Predicts spatial and time dependent currents and voltages
in part or whole-body biomodels
Numerous diagnostic and therapeutic applications, e.g.,
neurogenesis, cancer treatment, etc.
Fast parallelized computational approach
Simulation Models
From electrical standpoint, tissues are characterized
as conductivities and permittivities
Whole-body discretized within a cubic space
simulation volume
Cartesian grid of points along the three axes. Thus, at
most a total of six nearest neighbors
* Dimensions in millimeters
Numerical Models
Kirchhoff’s node analysis
[( A / L)d{V } / dt {V }( A / L)] 0
Recast to compute matrix only once
[ M ][V |t dt V |t ] [ B(t )]
For large models, matrix inversion is
intractable
LU decomposition of the matrix
Numerical Models
Voltage: User-specified timedependent waveform
Impose boundary conditions
locally
Actual data for conductivity
and permittivity
Results in extremely sparse
(asymmetric) matrix
[M]
Red: Total elements in the matrix
Blue: Nonzero Values
The Landscape of Sparse Ax=b Solvers
Direct
A = LU
Nonsymmetric
Symmetric
positive
definite
More Robust
Iterative
y’ = Ay
More General
Pivoting
LU
GMRES,
QMR, …
Cholesky
Conjugate
gradient
More Robust
Less Storage
Source: John Gilbert, Sparse Matrix Days in MIT 18.337
LU Decomposition
Source: Florin Dobrian
LU Decomposition
Source: Florin Dobrian
Computational Complexity
100 X 100 X 10 nodes: ~75 GB of memory (8-B floating precision)
Sparse data structure: ~ 6 MB (in our case)
Sparse direct solver: SuperLU-DIST
– Xiaoye S. Li and James W. Dimmel, “SuperLU-DIST: A Scalable
Distributed-Memory Sparse Direct Solver for Unsymmetric Linear
Systems”, ACM Trans. Mathematical Software, June 2003, Volume 29,
Number 2, Pages 110-140.
Fill reducing orderings with Metis
– G. Karypis and V. Kumar, “A fast and high quality multilevel scheme for
partitioning irregular graphs”, SIAM Journal on Scientific Computing,
1999, Volume 20, Number 1.
Performance on compute clusters
Time in Seconds
144,000-node Rat Model
Blue: Average iteration time
Cyan: Factorization time
Output: Visualization with MATLAB
Potential Profile at a depth of 12mm
Output: Visualization with MATLAB
Simulated Potential Evolution
Along the Entire 51-mm Width of the Rat Model
Deployment on
Mileva: 4-node cluster dedicated for SURAgrid purposes
Authentication
– ODU Root CA
– Cross certification with SURA Bridge
– Compatibility of accounts for ODU users
Authorization & Accounting
Initial Goals:
– Develop larger whole-body models with greater resolution
– Scalability tests
Grid Workflow
Establish user accounts for ODU users
– SURAgrid Central User Authentication and
Authorization System
– Off-line/Customized (e.g., USC)
Manually launch jobs based on remote resource
– SSH/GSISSH/SURAgrid Portal
– PBS/LSF/SGE
Transfer files
– SCP/GSISCP/SURAgrid Portal
Conclusions
Science:
– Electrostimulation has variety of diagnostic and
therapeutic applications
– While numerical simulations provide many advantages
over real experiments, they can be very arduous
Grid enabling:
– New possibilities with grid computing
– Grid-enabling an application is complex and time
consuming
– Security is nontrivial
Future Steps
Grid-enabling BioSim
– Explore alternatives for grid enabling BioSim
– Establish new collaborations
– Scalability experiments with large compute clusters
accessible via SURAgrid
Future applications:
– Molecular and Cellular Dynamics
– Computational Nano-Electronics
– Tools: Gromacs, DL-POLY, LAMMPS
References and Contacts
A Mishra, R Joshi, K Schoenbach and C Clark, “A Fast
Parallelized Computational Approach Based on Sparse LU
Factorization for Predictions of Spatial and Time-Dependent
Currents and Voltages in Full-Body Biomodels”, IEEE Trans.
Plasma Science, August 2006, Volume 34, Number 4.
http://www.lions.odu.edu/~rjoshi/
Ravindra Joshi, Ashutosh Mishra, Mike Sachon, Mahantesh
Halappanavar
– (rjoshi, amishra, msachon, mhalappa)@odu.edu
UAB and SURAgrid
Background
UAB's approach to grid deployment
Specific Testbed Application
UAB and SURAgrid Background
The University of Alabama at Birmingham (UAB) is
leading medical research campus in the South East
UAB was a participant in the National Science
foundation Middleware Initiative Test-bed (NMI Testbed) in 2002-2004
Evaluation of grid technologies in NMI Test-bed
sparked effort to build a campus grid: UABgrid
Collaboration with NMI Test-bed team members
leveraged synergistic interest in grid deployment
Managing Grid Middleware at
UAB
Over 10 years experience operating an
authoritative campus identity management
system
All members of UAB community have a selfselected network identity that is used as an
authoritative identity for human resource
records, desktop systems, and many other
systems on campus
Desire to leverage the same identity
infrastructure for grid systems
UABgrid Identity Management
Leverage Pubcookie as WebSSO system for webbased interface to authoritative, LDAP-based identity
management system
Create UABgrid CA to transparently assign grid
credentials using web based interface
Establish access to UABgrid resources based on
UABgrid PKI identity
Establish access to SURAgrid resources base on
cross certification of UABgrid CA with SURAgrid
Bridge CA
UABgrid Workflow
Establish grid PKI credentials for UAB users
– UAB Campus Identity Management System
– UABgrid CA cross certified with SURAgrid Bridge CA
Launch jobs via application specific web interface
– User authenticates via campus WebSSO
– Transparent initialization of grid credentials
– Submits job request
Collects results via application's web interface
Engaging Campus Researchers
Empower campus research applications with
grid technologies
Hide details of grid middleware operation
Select an application specific to the needs
campus researches
BLAST
Basic Local Alignment Search Tool (BLAST)
is an established sequence analysis tool that
performs similarity searches by calculating
statistical significance between a short query
sequence and a large database of
infrequently changing information such as
DNA and amino acid sequences
Used by numerous scientists in gene
discovery, categorization, and simulation.
BLAST algorithms
With the rapid development in sequencing
technology of large genomes for several
species, the sequence databases have been
growing at exponential rates
Parallel techniques for speeding up BLAST
searches:
– Database splitting BLAST (mpiBLAST,
TurboBLAST, psiBLAST)
– Query splitting BLAST (SS-Wrapper)
Goals of Grid Applications
Use distributed, dynamic, heterogeneous and
unreliable resources without direct user
intervention
Adapt to environment changes
Provide high level of usability while at least
maintaining performance levels
Dynamic BLAST goals
Complement BLAST with the power of the
grid
Master-worker application allowing for
dynamic load balancing and variable
resource availability based on Grid
middleware
Focuses on using small, distributed, readily
available resources (cycle harvesting)
Works with existing BLAST algorithm(s)
Dynamic BLAST architecture
Web interface
Dynamic BLAST
GridDeploy
GridWay
GT4
GT4
GT4
SGE
PBS
N/A
BLAST
BLAST
BLAST
MySQL
Perl
Java
scripts
Dynamic BLAST workflow
Jobs
Data analysis
Post-processing
File Parsing and
Fragment
creation
Job Monitor
GJ
Submit job
GJ
GJ
Dynamic BLAST performance
• Comparison of execution time
between query splitting BLAST,
Dynamic BLAST and mpiBLAST
for searching 1,000 queries against
yeast.nt database
QS-BLAST
mpiBLAST
Dynamic BLAST
0
20
40
60
80
100
120
140
• Comparison of execution time
of Dynamic BLAST on variable
number of nodes for 1,000 queries
against yeast.nt database
Time (in seconds)
Time (in seconds)
140
120
100
80
60
40
20
0
8 nodes
16 nodes
26 nodes
160
Deployment on SURAgrid
Incorporate ODU resource Mileva: 4-node cluster
dedicated for SURAgrid purposes
Local UAB BLAST web interface
– Leverage existing UAB Identity Infrastructure
– Cross certification with SURA Bridge CA
Authorization on a per-user basis
Initial Goals:
– Solidify the execution requirements of Dynamic BLAST
– Perform scalability tests
– Engage researchers further in the promise of grid computing
SURAgrid Experience
Combine Local needs with regional needs
No Need to Build Campus Grid in Isolation
Synergistic Collaborations
Leads to Solid Architectural Foundations
Leads to Solid Documentation
Extends Practical Experience for UAB
Students
Next Steps
Improve availability of web interface to non-UAB
users
– Leverage Shibboleth and GridShib to federate web interface
– Conversation begun on sharing technology foundation with
SURAgrid
Extend the number of resources running BLAST
– ODU integration helps document steps and is model for
additional sites
Explore other applications relevant to UAB research
community
References and Contacts
Purushotham Bangalore, Computer and
Information Sciences, UAB puri@uab.edu
Enis Afgan, Computer and Information
Sciences, UAB afgane@uab.edu
John-Paul Robinson, HPC Services, UAB IT
jpr@uab.edu
http://uabgrid.uab.edu/dynamicblast
SURAgrid Application Demos
Stop by the demo floor for a closer look at these
applications and more! Application demo team:
Mahantesh Halappanavar (ODU)
Sarat Sreepathi (NCSU)
Purushotham Bangalore (UAB)
Enis Afgan (UAB)
John-Paul Robinson (UAB)
Howard Lander (RENCI)
Victor Bolet (GSU)
Art Vandenberg (GSU)
Kate Barzee (SURA)
Mary Fran Yafchak (SURA)
Questions or comments?
For more information, or to join SURAgrid:
http://www.sura.org/SURAgrid
maryfran@sura.org