Chapter 23 Lecture PowerPoint - McGraw Hill Higher Education
advertisement

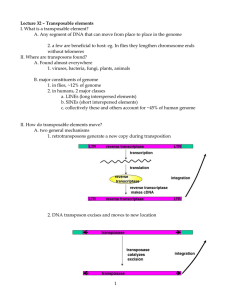
Lecture PowerPoint to accompany Molecular Biology Fifth Edition Robert F. Weaver Chapter 23 Transposition Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. 23.1 Bacterial Transposons • A transposable element moves from one DNA address to another • Originally discovered in maize, transposons have been found in all kinds of organisms – Bacteria – Plants – Humans 23-2 Discovery of Bacterial Transposons • • • • • Phage coat is made of protein Always has the same volume DNA is much denser than protein More DNA in phage, denser phage Extra DNAs that can inactivate a gene by inserting into the gene were the first transposons discovered in bacteria • These transposons are called insertion sequences (ISs) 23-3 Insertion Sequences • Insertion sequences are the simplest type of bacterial transposon • They contain only the elements necessary for their own transposition – Short inverted repeats at their ends – At least 2 genes coding for an enzyme, transposase that carries out transposition • Transposition involves: – Duplication of a short sequence in the target DNA – One copy of this sequence flanks the insertion sequence on each side after transposition 23-4 Generating Host DNA Direct Repeats 23-5 Complex Transposons • The term “selfish DNA” implies that insertion sequences and other transposons replicate at the expense of their hosts, providing no value in return • Some transposons do carry genes that are valuable to their hosts, antibiotic resistance is among the most familiar 23-6 Antibiotic Resistance and Transposons • Donor plasmid has Kanr, harboring transposon Tn3 with Ampr • Target plasmid has Tetr • After transposition, Tn3 has replicated and there is a copy in target plasmid • Target plasmid now confers both Ampr, Tetr 23-7 Transposition Mechanisms • Transposons are sometimes called “jumping genes”, DNA doesn’t always leave one place for another • When it does, nonreplicative transposition – “Cut and paste” – Both strands of original DNA move together from 1 place to another without replicating • Transposition frequently involves DNA replication – – – – 1 copy remains at original site New copy inserts at the new site Replicative transposition “Copy and paste” 23-8 Replicative Transposition of Tn3 • In first step, 2 plasmids fuse, phage replication, forms a cointegrate – coupled through pair of Tn3 copies • Next is resolution of cointegrate, breaks down into 2 independent plasmids, catalyzed by resolvase gene product 23-9 Detailed Tn3 Transposition 23-10 Nonreplicative Transposition • Starts with same 2 first steps as in replicative transposition • New nicks occur at arrow marks • Nicks liberate donor plasmid minus the transposon • Filling gaps and sealing nicks completes target plasmid and its new transposon 23-11 23.2 Eukaryotic Transposons • Transposons have powerful selective forces on their side • Transposons carry genes that are an advantage to their hosts – Their host can multiply at the expense of completing organisms – Can multiply the transposons along with rest of their DNA • If transposons do not have host advantage, can replicate themselves within their hosts 23-12 Examples of Transposable Elements • Variegation in the color of maize kernels is caused by multiple reversions of an unstable mutation in the C locus, responsible for kernel color • Mutation and its reversion result from Ds (dissociation) element – Transposes into the C gene – Mutates it – Transposes out again, revert to wild type 23-13 Ds and Ac of Maize • Ds cannot transpose on its own • Must have help from an autonomous transposon, Ac (for activator) – Ac supplies transposase – Ds is an Ac element with most of its middle removed – Ds needs • A pair of inverted terminal repeats • Adjacent short sequences that Ac transposase can recognize 23-14 Transposable Elements in Maize 23-15 Structures of Ac and Ds 23-16 P Elements • The P-M system of hybrid dysgenesis in Drosophila is caused by conjunction of 2 factors: – Transposable element (P) contributed by the male – M cytoplasm contributed by the female allows transposition of the P element • Hybrid offspring of P males and M females suffer multiple transpositions of P element • Damaging chromosomal mutations are caused that render the hybrids sterile • P elements have practical value as mutagenic and transforming agents in genetic experiments with Drosophila 23-17 23.3 Rearrangement of Immunoglobulin Genes • Mammalian genes use a process that closely resembles transposition for: – B cell receptors (antibodies when secreted) – T cell receptors • Recombinases involved in these processes have similar structures 23-18 Antibody Structure • Antibody is composed of 4 polypeptides – 2 heavy chains – 2 light chains • Sites called variable regions – Vary from 1 antibody to another – Gives proteins their specificity • Rest of protein is constant region 23-19 Immune System Diversity • Enormous diversity of immune system is generated by 3 basic mechanisms: – Assembly of genes for antibody light chains and heavy chains from 2 or 3 component parts – Joining the gene segments by an imprecise mechanism that can delete bases or add extra bases – A high rate of somatic mutations, during proliferation of a clone of immune cells (this only applies to the B cell receptor/antibody) 23-20 Rearrangement of Antibody Light Chain Gene 23-21 Antibody Heavy Chain Coding Regions Human heavy chain is encoded in – – – – 48 variable segments 23 diversity segments 6 joining segments 1 constant segment 23-22 Recombination Signals • The recombination signal sequences (RSSs) in V(D)J recombination consist of: – Heptamer – Nonamer – Separated by 12-bp or 23-bp spacers • Recombination occurs only between a 12 signal and a 23 signal (12/23 rule) • Guarantees that only 1 of each coding region is incorporated into the rearranged gene 23-23 Signals for V(D)J joining 23-24 The Recombinase • Recombination-activating gene (RAG-1) stimulated V(D)J joining activity in vivo • Another gene tightly liked to RAG-1 also works in V(D)J joining, RAG-2 • These genes, RAG-1 and RAG-2, are expressed only in pre-B and pre-T cells 23-25 Mechanism of V(D)J Recombination • RAG1 and RAG2 introduce single-strand nicks into DNA adjacent to either a 12 signal or 23 signal • Results in transesterification where newly created 3’-OH group: – Attacks the opposite strand – Breaks it – Forms hairpin at the end of the coding segment • Hairpins then break in an imprecise way that allows joining of coding regions with loss of bases or gain of extra bases 23-26 23.4 Retrotransposons • Retrotransposons replicate through an RNA intermediate • Retrotransposons resemble retroviruses – Retroviruses can cause tumors in vertebrates – Some retroviruses cause diseases such as AIDS • Before studying retrotransposons, look at replication of the retroviruses 23-27 Retroviruses • Class of virus is named for its ability to make a DNA copy of its RNA genome • This reaction is the reverse of the transcription reaction – reverse transcription • Virus particles contain an enzyme that catalyzes reverse transcription reaction, called reverse transcriptase 23-28 Retrovirus Replication • Viral genome is RNA, with long terminal repeats at each end • Reverse transcriptase makes linear, ds-DNA copy of RNA • ds-DNA copy integrates back into host DNA = provirus • Host RNA polymerase II transcribes the provirus into genomic RNA • Viral RNA packaged into a virus particle 23-29 Model for Synthesis of Provirus DNA • RNase H degrades the RNA within the RNA-DNA hybrids created during the replication process • Host tRNA serves as primer for reverse transcriptase • Finished ds-DNA copy of viral RNA is then inserted into the host genome • It can be transcribed by host polymerase II along with the host genome 23-30 Retrotransposons • Several eukaryotic transposons transpose in a way similar to retroviruses – Ty of yeast – copia of Drosophila • Start with DNA in the host genome – Make an RNA copy – Reverse transcribe it within a virus-like particle into DNA that can insert into new location • HERVs likely transposed in the same way until the ability to transpose was lost – HERV = human endogenous retroviruses 23-31 Model for Ty Transposition 23-32 Non-LTR Retrotransposons • LTR are lacking in most retrotransposons • Most abundant type lacking LTR are LINEs and LINE-like elements – Long interspersed elements – Encode an endonuclease that nicks target DNA – Takes advantage of new DNA 3’-end to prime reverse transcriptase of element RNA – After 2nd strand synthesis, element has been replicated at target site • New round of transposition begins when the LINE is transcribed • LINE polyadenylation signal is weak, so transcription of a LINE often includes exons of downstream host DNA 23-33 Nonautonomous Retrotransposons • Nonautonomous retrotransposons include very abundant human Alu elements and similar elements in other vertebrates • Cannot transpose by themselves as they do not encode any proteins • Take advantage of retrotransposition machinery of other elements such as LINE • Processed pseudogenes likely arose in same manner 23-34 Group II Introns • Group II introns – Retrohome to intronless copies same gene by: • Insertion of an RNA intron into the gene • Followed by reverse transcription • Then second-strand synthesis – Retrotranspose by: • Insertion of an RNA intron into an unrelated gene • Target-primed reverse transcription • Lagging-strand DNA fragments as primers • Group II retrotransposition: – Forerunner of eukaryotic spliceosomal introns – Accounted for appearance in higher eukaryotes 23-35 Retrohoming 23-36