King
advertisement
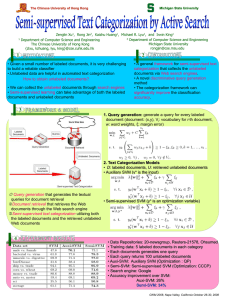
Semi-supervised learning for protein classification Brian R. King Chittibabu Guda, Ph.D. Department of Computer Science University at Albany, SUNY Gen*NY*sis Center for Excellence in Cancer Genomics University at Albany, SUNY 1 The problem Develop computational models of characteristics of protein structure and function from sequence alone using machine-learned classifiers Input: Data Output: A model (function) h : X Y Traditional approach: supervised learning Challenges: Experimentally determined data – Expensive, limited, subject to noise/error Large repositories of unannotated data Data representation, bias from unbalanced / underrepresented classes, etc. Swiss-Prot 54.5: 289,473 TrEMBL 37.5: 5,035,267 AIM: Develop a method to use labeled and unlabeled data, while improving performance given the challenges presented by small, unbalanced data 2 Solution Semi-supervised learning Use Dl and Du for model induction Method: Generative, Bayesian probabilistic model Based on ngLOC – supervised, Naïve Bayes classification method Input / Feature Representation: Sequence n-gram model Assumption – multinomial distribution Use EXPECTATION MAXIMIZATION! Test setup Prediction of subcellular localization Eukaryotic, non-plant sequences only Dl : Data annotated with subcellular localization for eukaryotic, non-plant sequences IID – Sequence and n-grams DL-2 – EXT/PLA (~5500 sequences, balanced) DL-3 – GOL [65%] / LYS [14%] /POX [21%] (~600 sequences, unbalanced) Du : Set from ~75K eukaryotic, non-plant protein sequences. Comparative method Transductive SVM 3 Algorithms based on EM 0.9 No Unlabeled Sequences 25,000 Unlabeled Sequences # Unlabeled 0 2,000 4,000 6,000 8,000 50,000 Macro-averaged F1 0.8 0.7 0.6 0.5 0.4 0.3 0 1000 2000 TSVM macF1 Accuracy 0.499 0.626 0.641 0.653 0.645 0.661 0.659 0.667 NA NA NA NA 3000 4000 5000 Number of Labeled Protein Sequences Basic EM on DL-2 Varied labeled data 25,000 UL sequences Most improvement when data is limited 6000 EM-λ macF1 Accuracy 0.561 0.728 0.602 0.735 0.633 0.743 0.645 0.743 0.672 0.747 0.677 0.752 EM-λ on DL-3 data λ – controls effect of UL data on parameter adjustments ALL labeled data (~600) Varied UL data EM- λ outperforms TSVM on this problem (Failed to converge on large amounts of UL data, despite parameter selection) NOTE – TSVM performed very well on binary, balanced classification problems 4 Algorithm – EM-CS Core ngLOC method outputs a confidence score (CS) Improve running time through intelligent selection of unlabeled instances CS(xi) > CSthresh? Use the instance Test on DL-3 data: CSthresh Precision Percentile 40 100% 94 38.25 97% 87 37 88% 71 36 85% 50 35 78% 21 First, determine range of CS scores through cross-validation without UL: 33.5-47.8 (Dependent on level of similarity in data, size of dataset.) CSthresh 0 35 36 37 38.25 40 UL data 0 60,733 6814 3810 2388 1474 726 macF1 0.561 0.650 0.662 0.674 0.676 0.672 0.621 Accuracy 0.728 0.690 0.721 0.735 0.741 0.752 0.740 Using only sequences that meet or exceed CSthresh significantly reduces UL data required (97.5% eliminated) NOTE: it is possible to reduce UL data too much. 5 Conclusion Benefits: Probabilistic Extract unlabeled sequences of “high-confidence” Difficult with SVM or TSVM Extraction of knowledge from model Discriminative n-grams and anomalies Time: Significantly lower than SVM and TSVM Space: Dependent on n-gram model Can use large amounts of unlabeled data Applicable toward prediction of any structural or functional characteristic Outputs a global model Again, difficult with SVM or TSVM Computational resources Information theoretic measures, KL-divergence, etc. Transduction is not global! Most substantial gain with limited labeled data Current work in progress: TSVMs Improve performance on smaller, unbalanced data Select an improved smaller dimensional feature space representation Ensemble classifiers, Bayesian model averaging, Mixture of experts 6