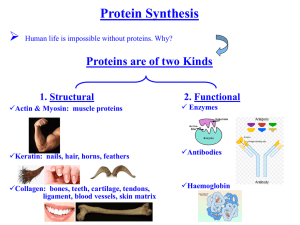
File - Genetics/Genomics Competency Center
advertisement

Gene Expression Overview By Salwa Hassan Teama M.D. N.C.I. Cairo university 1 Gene Expression Eukaryotic Cell The Gene Structure Protein Synthesis Prokaryotes Vs Eukaryotes 2 Gene Expression Gene expression is the process by which a genes information is converted into the structures and functions of a cell by a process of producing a biologically functional molecule of either protein or RNA (gene product) is made. Gene expression is assumed to be controlled at various points in the sequence leading to protein synthesis. 3 Eukaryotic Cell 4 Gene Structure Eukaryotic gene structure: Most eukaryotic genes in contrast to typical bacterial genes, the coding sequence (exons) are interrupted by noncoding DNA (introns). The gene must have ( Exon; start signals; stop signals; regulatory control elements). 5 Protein Synthesis Protein Synthesis is the process in which cells build proteins from information in a DNA gene in a two major steps: I-Transcription and II-Translation Transcription : synthesis of an RNA (mRNA) that is complementary to one of the strands of DNA. Translation : ribosomes read a messenger RNA and make protein according to its instruction. 6 Protein Synthesis 7 Transcription 8 http://biology.unm.edu/ccouncil/Biology_124/Images/transcription.gif Transcription RNA polymerase copies both the exons and the introns. The stretch of DNA that is transcribed into an RNA molecule is called a transcription unit. A transcription unit that is translated into protein contains coding sequence that is translated into protein and sequences that direct and regulate protein synthesis; Transcription proceeds in the 5' → 3' direction. Transcription is divided into 3 phases: Initiation, Elongation and Termination.. 9 RNA polymerase; eukaryotic nuclei contain three RNA polymerases . RNA polymerase I is found in the nucleolus; the other two polymerases are located in the nucleoplasm. The three nuclear RNA polymerase have different roles in transcription. Polymerase I makes a large precursor to the major rRNA (5.8S,18S and 28S rRNA in vertebrates). Polymerase II synthesizes hnRNAs, which are precursors to mRNAs. It also make most small nuclear RNAs (snRNAs). Polymerase III makes the precursor to 5SrRNA, the tRNAs and several other small cellular and viral RNAs. 10 Initiation The general transcription factors combine with RNA polymerase to form a preinitiation complex that is competent to initiate transcription as soon as nucleotide are available. 11 Initiation The enzyme RNA polymerase recognizes a promoter, which lies upstream of the gene. The polymerase binding causes the unwinding of the DNA double helix. This is followed by initiation of RNA synthesis at the starting point. The RNA polymerase starts building the RNA chain, it assembles ribonucleotides triphosphates: ATP; GTP; CTP and UTP into a strand of RNA. After the first nucleotide is in place, the polymerase joins a second nucleotide to the first, forming the initial phosphodiester bond in the RNA chain. 12 Elongation RNA polymerase directs the sequential binding of riboncleotides to the growing RNA chain in the 5`-3` direction. Each ribonucleotide is inserted into the growing RNA strand following the rules of base pairing. This process is repeated till the desired RNA length is synthesized……………………. 13 Termination Other regions at the end of genes; called terminators, signal termination. These work in conjunction with RNA polymerase to loosen the association between RNA product and DNA template. The result is that the RNA dissociate from RNA polymerase and DNA and so stop transcription. The product is immature RNA or pre mRNA (Primary transcript). 14 RNA Processing Pre-mRNA → mRNA Capping: Synthesis of the cap. The 5` cap is a 7- methylguanosine (m7G) . The cap protects the mRNA from being degraded by enzymes; enhancement of mRNA translatability. Splicing: Step-by-step removal of introns present in the pre-mRNA and joining of the remaining exons. The removal of introns and joining of exons takes place on a special structures called spliceosomes. Polyadenylation: Synthesis of the poly (A) tail involves cleavage of its 3' end and then the addition of about 200 adenine residues to form a poly (A) tail; This completes the mRNA molecule (mature mRNA), which is now ready for export to the cytosol for protein synthesis. 15 RNA Processing 16 RNA Splicing 17 Alternative Splicing Alternative splicing: is a very common phenomenon in higher eukaryotes. It is a way to get more than one protein product out of the same gene and a way to control gene expression in cells. 18 Translation http://www.nature.com/embor/journal/v4/n9/images/embor923-f3.jpg 19 Translation Translation is the process by which ribosomes read the genetic message in the mRNA and produce a protein product according to the message's instruction. 20 Translation Requirement for translation Ribosomes tRNA mRNA template Amino Acids Initiation factors Elongation factors Termination factors Aminoacyl tRNA synthetase enzymes Energy source 21 Ribosomes are the site of protein biosynthesis using the mRNA as a template, the ribosome traverses each codon of the mRNA, pairing it with the appropriate amino acid. This is done using molecules of transfer RNA (tRNA) containing a complementary anticodon on one end and the appropriate amino acid on the other. http://www.molecularexpressions.com/cells/ribosomes/images/ribosomesfigure1.jpg 22 tRNA Act as adaptors that can bind an amino acid at one end and interact with the mRNA at the other. mRNA Source of coding information for the protein synthesis system. Contains start and stop signals for translation. Eukaryotic mRNA is capped. This is used as the recognition feature for ribosome binding. The site at which protein synthesis begins on the mRNA is especially crucial, since it sets the reading frame for the whole length of the message. An error of one nucleotide either way at this stage would cause every subsequent codon in the message to be misread, so that a nonfunctional protein would result, the rate of initiation thus determines the rate at which the protein is synthesized. 23 Amino acids are the monomers which are polymerized to produce proteins. The amino acids are loaded onto tRNA molecules for use in the process of translation. Initiation factors help the ribosome, initiator tRNA, and other components assemble the at the correct location on the mRNA and ensure that protein synthesis starts in the correct reading frame . Elongation factors are responsible for moving the ribosome along the mRNA and maintain the correct reading frame. Facilitate removal of "used" tRNAs and bringing in "new" tRNAs. Termination factors recognize the stop codons and release proteins and ribosomes. Aminoacyl tRNA synthetase enzymes: It catalyze the covalent attachment of an amino acids to the end of the corresponding tRNA. Energy source: ATP or GTP which are synthesized in the mitochondria. 24 Translation Preparatory steps for protein synthesis: First, aminoacyl tRNA synthetase join amino acid to their specific tRNA. Second, ribosomes must dissociate into subunits at the end of each round of translation. The protein synthesis occur in 3 phases: 1- Accurate and efficient initiation occurs, the ribosomes binds to the mRNA, and the first amino acid attached to its tRNA. 2- Chain elongation, the ribosomes adds one amino acid at a time to the growing polypepyide chain. 3- Accurate and efficient termination, the ribosomes releases the mRNA and the polypeptide. 25 Translation: Initiation The initiation phase of protein synthesis requires over 10 eukaryotic Initiation Factors (eIFs): Factors are needed to recognize the cap at the 5` end of an mRNA and binding to the 40s ribosomal subunit. Binding the initiator Met-tRNAiMet (methionyl- tRNA) to the 40S small subunit of the ribosome. Scanning to find the start codon by binding to the 5` cap of the mRNA and scanning downstream until they find the first AUG (initiation codon). The start codon must be located and positioned correctly in the P site of the ribosome and the initiator tRNA must be positioned correctly in the same site. Once the mRNA and initiator tRNA are correctly bound, the 60S large subunit binds to form 80 s initiation complex with release of the eIF factors. 26 The large ribosomal subunit contains three tRNA binding sites, designated A, P, and E. The A site binds an aminoacyl-tRNA (a tRNA bound to an amino acid); the P site binds a peptidyl-tRNA (a tRNA bound to the peptide being synthesized); and the E site binds a free tRNA before it exits the ribosome. 27 Elongation Transfer of proper aminoacyl-tRNA from cytoplasm to A-site of ribosome; Peptide bond formation; Peptidyl transferase forms a peptide bonds between the amino acid in the P site and the newly arrived aminoacyl tRNA in the A site. This lengthens the peptide by one amino acids. 28 Elongation Translocation; translocation of the new peptidyl t-RNA with its mRNA codon in the A site into the free P site occurs Now the A site is free for another cycle of aminoacyl t-RNA codon recognition and elongation. Each translocation events moves mRNA , one codon length through the ribosomes. 29 Termination Translational termination requires specific protein factors identified as releasing factors, RFs in E. coli and eRFs in eukaryotes. The signals for termination are the same in both prokaryotes and eukaryotes. These signals are termination codons present in the mRNA. There are 3 termination codons, UAG, UAA and UGA. 30 Termination After multiple cycles of elongation and polymerization of specific amino acids into protein molecules, a nonsense codon = termination codon of mRNA appear in the A site. The is recognized as terminal signal by eukaryotic releasing factors (eRF) which cause the release of the newly synthesized protein from the ribosomal complex. 31 Protein Synthesis http://bioinfo.bact.wisc.edu/themicrobialworld/lysozyme.gif 32 Reading the instruction means translating the code in the RNA from bases (building block of DNA and RNA) to amino acids (building block of Protein proteins). Synthesis www.bseinquiry.gov.uk/report/volume2/fig1_2.htm 33 Prokaryotic vs. Eukaryotic Eukaryotic DNA is wound around histones to form nucleosomes and packaged as chromatin. Chromatin has a strong influence on the accessibility of the DNA to transcription factors and the transcriptional machinery including RNA polymerase. Eukaryote genes are not grouped in operons. each eukaryote gene is transcribed separately, with separate transcriptional controls on each gene. Protein synthesis takes place in the cytoplasm while transcription and RNA processing take place in the nucleus. Essentially all humans' genes contain introns. A notable exception is the histone genes which are intronless. 34 Prokaryotic vs. Eukaryotic Eukaryotic mRNA is modified through RNA splicing. Eukaryotic mRNA is generally monogenic (monocistronic); code for only one polypeptide. Eukaryotes have a separate RNA polymerase for each type of RNA. Eukaryotic mRNA contain no Shine-Dalgarno sequence to show the ribosomes where to start translating. Instead, most eukaryotic mRNA have caps at their 5` end which directs initiation factors to bind and begin searching for an initiation codon. Eukaryotic protein synthesis initiation begins with methionine not N formyl- methionine. In eukaryotes, polysomes are found in the cytoplasm. 35 Prokaryotic vs. Eukaryotic Bacterial genetics are different. Prokaryote genes are grouped in operons. Prokaryotes have one type of RNA polymerase for all types of RNA, mRNA is not modified The existence of introns in prokaryotes is extremely rare. To initiate transcription in bacteria, sigma factors bind to RNA polymerases. RNA polymerases/ sigma factors complex can then bind to promoter about 40 deoxyribonucleotide bases prior to the coding region of the gene. In prokaryotes, the newly synthesized mRNA is polycistronic (polygenic) (code for more than one polypeptide chain). In prokaryotes, transcription of a gene and translation of the resulting mRNA occur simultaneously. So many 36 polysomes are found associated with an active gene. References & Further Reading Robert F.Weaver. Molecular Biology. Fourth Edition. Page 600. McGraw-Hill International Edition. ISBN 978-0-07-110216-2 Innis,David H. Gelfand,John J. Sninsky PCR Applications: Protocols for Functional Genomics: ISBN:0123721865 Daniel H. Farkas. DNA Simplified: The Hitchhiker's Guide to DNA. Washington, DC: AACC Press, 1996, ISBN 0915274-84-1. William B. Coleman,Gregory J. Tsongalis: Molecular Diagnostics: For the Clinical Laboratorian: ISBN 1588293564... Robert F. Mueller,Ian D. Young. Emery's Elements of Medical Genetics: ISBN. 044307125X Daniel P. Stites,Abba T. Terr. Basic Human Immunology: ISBN. 0838505430 Bruce Alberts, Alexander Johnson, Julian Lewis, Martin Raff, Keith Roberts, and Peter Walter. Molecular Biology of the cell. ISBN. 9780815341055 http://www.pubmedcentral.nih.gov/ www.medscape.com www.ebi.ac.uk/2can good introduction to bioinformatics and molecular biology http://www.genomicglossaries.com http://www.gene.ucl.ac.uk/nomenclature/guidelines.html defines the nomenclature for human genes http://www.accessexcellence.org http://users.rcn.com/jkimball.ma.ultranet/BiologyPages/C/Codons.html http://www.web-books.com/MoBio/ http://www.expasy.org http://www.emc.maricopa.edu/faculty/farabee/BIOBK/BioBookPROTSYn.html Cell & Molecular Biology online: http://www.cellbio.com/recommend.html http://www.ornl.gov/sci/techresources/Human_Genome/glossary/glossary.shtml%20 http://www.genome.gov/10000715 http://www.ncbi.nlm.nih.gov/About/primer/mapping.html http://www.lilly.com/research/discovering/targets.html http://www.informatics.jax.org/expression.shtml www.wikipdia.com http://www.biology.arizona.edu/cell_bio/tutorials/pev/page2.html http://www.genome.ou.edu/protocol_book/protocol_index.html 37 38