Topic 8: Quantitative Genetics
advertisement

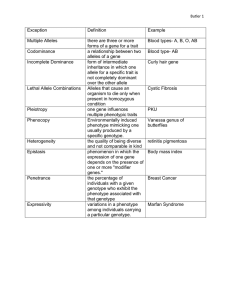
Quantitative genetics: traits controlled my many loci Key questions: what controls the rate of adaptation? Example: will organisms adapt to increasing temperatures or longer droughts fast enough to avoid extinction? What is the genetic basis of complex traits with continuous variation? Quantitative genetics vs. population genetics Population genetics Quantitative genetics Breeder’s equation Breeder’s equation: R = h2S Polygenic Inheritance Leads to a Quantitative Trait TRAIT A B C Z1 # of individuals LOCUS D Z E F S, the selection differential R, the selection response, Problems in predicting the evolution of quantitative traits - Dominance Epistasis Environmental effects Environment alters gene expression - epigenetics yellow: no difference; red or green = difference age 3 age 50 Overall: not all variation is heritable Major questions in quantitative genetics • How much phenotypic variation is due to genes, and how much to the environment? • How much of the genetic variation is due to genes of large effect, and how much to genes of small effect? Measuring heritability Variance: _ Vp= Σi (Xi – X)2 --------------(N – 1) Variance components VP = total phenotypic variance VP = VA + VD + VE + VGXE VA = Additive genetic variance VD = Dominance genetic variance (nonadditive – can include epistasis) VE = Variance among individuals experiencing different environments VGXE = Variance due to environmental variation that influences gene expression (not covered in text) Heritability = h2 = VA / VP The proportion of phenotypic variance due to additive genetic variance among individuals h2 = VA / (VA + VD + VE + VGXE) Heritability can be low due to: Additive vs. dominance variance Additive: heterozygote is intermediate Dominance: heterozygote is closer to one homozygote (Difference from line is due to dominance) Dominance and heritability Dominance Genotype AA AA’ A’A’ f(A) = p = 0.5 f(A’) = q = 0.5 Start in HWE f(AA) = 0.25 f(AA’) = 0.50 f(A’A’) = 0.25 Toe len (cm) 0.5 1.0 1.0 Codominant (additive) Genotype Toe len (cm) AA 0.5 AA’ 0.75 A’A’ 1.0 f(A) = p = 0.5 f(A’) = q = 0.5 Start in HWE f(AA) = 0.25 f(AA’) = 0.50 f(A’A’) = 0.25 Dominance and heritability II Dominance Codominant (additive) 1 1 0. 9 0. 9 0. 8 0. 8 0. 7 0. 7 Starting Genotype frequencies 0. 6 0. 5 0. 4 0. 3 0. 2 0. 1 0. 6 0. 5 0. 4 0. 3 0. 2 0. 1 0 0 AA AA' A 'A ' AA Genotype 1 1 0.9 0.9 0.8 0.8 0.7 0.6 0.5 Mean = 0.875 cm 0.7 0.6 Starting Phenotype frequencies 0.4 0.3 0.2 0.1 AA' Genotype A 'A ' Mean = 0.75 cm 0.5 0.4 0.3 0.2 0.1 0 0 0.5 0.75 1 Phenotype (toe len – cm) 0.5 0.75 1 Phenotype (toe len – cm) Select toe length = 1 cm Dominance Additive 1 1 0. 9 0. 9 0. 8 0. 8 0. 7 0. 7 Starting Genotype frequencies 0. 6 0. 5 0. 4 0. 3 0. 2 0. 1 0. 6 0. 5 0. 4 0. 3 0. 2 0. 1 0 0 AA AA' A 'A ' AA Genotype 1 1 0.9 0.9 0.8 0.8 0.7 0.6 0.5 Mean = 1.0 cm 0.7 0.6 Starting Phenotype frequencies 0.4 0.3 0.2 0.1 AA' Genotype A 'A ' Mean = 1.0 cm 0.5 0.4 S= 0.3 0.2 0.1 0 0 0.5 0.75 0.5 1 Phenotype (toe len – cm) S= 0.75 1 Phenotype (toe len – cm) Effects of dominance: genotypes after random mating Dominance Codominant (additive) 1 1 0. 9 0. 9 0. 8 0. 8 0. 7 0. 7 0. 6 Genotype freq after selection, before mating 0. 5 0. 4 0. 3 0. 2 0. 1 0. 6 0. 5 0. 4 0. 3 0. 2 0. 1 0 AA AA' 0 A 'A ' AA Genotype AA' Genotype A 'A ' 1 1 0. 9 0. 9 0. 8 0. 8 0. 7 0. 7 0. 6 0. 6 Genotype freq after mating 0. 5 0. 4 0. 3 0. 2 0. 5 0. 4 0. 3 0. 2 0. 1 0. 1 0 0 AA AA' A 'A ' AA Genotype AA' Genotype A 'A ' Effects of dominance: phenotypes after random mating Dominance Codominant (additive) 1 1 0.9 0.9 0.8 0.8 0.7 Phenotype freq after selection, before mating 0.6 0.5 0.4 0.3 0.2 0.1 0.7 0.6 0.5 0.4 0.3 0.2 0.1 0 0.5 0.75 Genotype 0 1 0.5 1 0.7 0.6 1 0.9 0.9 0.8 0.75 1 0.8 mean = 0.954 0.7 0.6 0.5 0.5 Phenotype freq after mating 0.4 0.3 0.2 mean = 1.0 0.4 0.3 R=1 - .75 0.2 0.1 0.1 0 0 0.5 0.75 Phenotype 1 0.5 R = 954 - .875 = 0.079 0.75 Phenotype 1 = 0.25 Effect of dominance on heritability Dominant S= R= R = h 2S h2 = R / S = Codominant (additive) S= R= R = h 2S h2 = Second problem predicting outcome of selection: epistasis Example: hair color in mammals MC1 agouti MC1receptor Agouti is antagonist for MC1R. If agouti binds, no dark pigment produced Second problem predicting outcome of selection: epistasis Epistasis MC1 agouti MC1receptor Normal receptor Mutant: never Mutant: always dark pigment dark pigment (yellow labs) Epistasis example Genotype Phenotype EE / AA dark tips, light band ee / -blond / gold / red Ed- / -all dark EE / Adblond / gold / red Want dark fur: population ee / AA Ee / AdA Ee / AA Epistasis example ii Cross: Ee / AdA genotype EE / AdAd EE / AdA EE / AA x Ee / AdA phenotype freq 1/16 1/8 1/16 Ee / AdAd Ee / AdA Ee / AA 1/8 1/4 1/8 ee / AdAd ee / AdA ee / AA 1/16 1/8 1/16 Measuring h2: Parent-offspring regression Estimating h2 Analysis of related individuals Measuring the response of a population, in the next generation, to selection Heritability is estimated as the slope of the least-squares regression line h2 data: Darwin’s finches h2 example: Darwin’s finches mean before selection: 9.4 mean after selection: 10.1 S = 10.1 - 9.4 = 0.7 h2 example: Darwin’s finches II mean before selection: 9.4 mean of offspring after selection: 9.7 Response to selection: 9.7 – 9.4 = 0.3 R = h2S; 0.3 = h2 * 0.7 h2 = R/S = 0.3 / 0.7 = 0.43 Controlling for environmental effects on beak size • song sparrows: cross fostering Offspring vs. biological parent (h2); vs. foster parent (VE) Smith and Dhondt (1980) Cross fostering in song sparrows Testing for environmental effects How can we determine the effect of the environment on the phenotype? Two genotypes in two environments: possible effects on phenotype Example of environmental effects: locusts Environmental effects: carpenter ant castes queen male major worker minor worker Effect of GxE: predicting outcome of selection 7 yarrow (Achillea millefolium) genotypes 2 gardens Clausen, Keck, and Heisey (1948) What happens to h2 when selection occurs? Modes of selection Gen. 2 Gen. 1 Gen. 0 freq. trait, z Mode of selection: directional freq. freq. trait, z trait, z Mode of selection: stabilizing freq. freq. trait, z trait, z Mode of selection: disruptive freq. freq. trait, z trait, z Disruptive selection Distribution of mandible widths in juveniles that died (shaded) and survived (black) Fitness Beak depth Beak depth Fitness • Darwin’s finches: beak width is correlated with beak depth Beak width Complications: correlations Beak width Detecting loci affecting quantitative traits (QTL) QTLs and genes of major effects How important are genes of major effect in adaptation? QTL analysis: Quantitative Trait Loci – where are the genes contributing to quantitative traits? • Approach – two lineages consistently differing for trait of interest (preferably inbred for homozygosity) – Identify genetic markers specific to each lineage (eg microsatellite markers) – make crosses to form F1 – generate F2s and measure trait of interest – test for association between markers and trait – Estimate the effect on the phenotype of each marker Example: Mimulus cardinalis and Mimulus lewisii Mimulus cardinalis Mimulus lewisii Locate lineage-specific markers Mimulus lewisii Mimulus cardinalis Microsatellite length 250 (M. l.) or 254 (M. c.) M. lewisii specific Legend M. cardinalis specific non-specific Cross lines QTL Mapping: crosses F1 F2 recombinants intermediate phenotype scrambled phenotypes QTL analysis: trait associations Homozygotes at marker 2 are closer to one parent Heterozygotes at marker 2 are intermediate in trait values Trait analysis For each marker, ask whether changing genotype affects phenotype QTL probabilities Estimate the probability of location of QTL based on association of markers and recombination probabilities (eg CD34B rarely associated with pH, CD34A nearly always, TG63 rarely) Markers Example study: basis of floral traits in two Mimulus species (Schemske and Bradshaw, PNAS 1999) Pollination syndrome changes during the evolution of the Mimulus group: hummingbird or bee F2 plants showed variation for most floral traits F1 M. lewisii F2 plants M. cardinalis Most traits had multiple QTL but one explaining > 25% of variation Few genes of large effect important in this case Case study: domestication traits in sunflowers • Most traits showed many genes of small effect (< 10%) • Problems: gene map resolution is low – 35,000 genes per plant genome – 100 markers on genetic map: 350 genes per marker • A “weak” QTL can be due to – nearby gene of weak effect – more distant gene of strong effect (looks weak due to recombination between marker and locus) Recap: quantitative genetics • Are traits heritable? – Usually find heritability – However, environmental effects can be large • Are genes controlling quantitative traits of large effect or small effect? – Some important genes for adaptation of large effect. – Overall pattern still unclear Questions 1. 2. 3. Human height is highly heritable: among university students in the US, the heritability is 0.84. Yet, during the 1980s, when Guatemalan refugees fled the civil war to the United States, 12 year old Mayan children were four inches taller in the United States than in Guatemala. How is this possible? Consider the following scenario: In October 2006 you banded and weighed all of the adult burrowing owls near Osoyoos. The average owl weighed 207.8 gm. The winter of 2006-2007 was especially cold, and many owls died before they bred in the spring. When you weighed those owls that survived to breed in April of 2007, their average weight was 211.8 gm. If the heritability of owl body weight is 0.25, what is the expected mean body weight in the NEXT generation of adults, assuming none die before they become adults? Data demonstrating stabilizing selection for human head size at birth were collected in 1951 in the United States. If you were to collect the same data now, would you expect to find the same pattern? Why or why not? Would it matter where in the world you did your study? Also, see posted quantitative genetics practice questions for further practice.