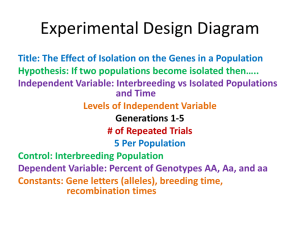
Tests for Association

Epidemiology 719
Quantitative methods in genetic epidemiology
Bhramar Mukherjee and Sebastian Zoellner bhramar@umich.edu
szoellne@umich.edu
Acknowledgements
• Peter Kraft (HSPH)
• Ken Rice (UW)
• Nilanjan Chatterjee (NCI)
• Stephen Channock (NCI)
• Lu Wang (UM)
• Nan Laird (HSPH)
• Goncalo Abecasis (UM)
A brave new world
Course Overview
Reverse Effects
Central Course Theme
Genetic Association and
Gene-Environment Interaction
Course Advice for You:
Assigned Paper 1
Assigned Paper 1
• GWAS of Age-related macular degeneration
• Initial GWAS identified four loci explaining one-half of the heritability. Appreciable predictive power.
• Additional GWAS to explain remaining heritability. Combined scan vs replication.
Meta-Analysis.
Assigned Paper 2
Assigned Paper 2
• Collaborative Association Study of
Psoriasis
• Examined ~1,500 cases / ~1,500 controls at ~500,000 SNPs
• • Examined 20 promising SNPs in extra
~5,000 cases / ~5,000 controls
• Outcome: 7 regions of confirmed association with psoriasis
Assigned Paper 3
Assigned Paper 3
• Meta-analysis of colorectal cancer
(COGENT study) .
• A thorough evaluation of ten confirmed loci for colorectal cancer. Very detailed.
Supplementary material also available online.
• Interesting combination of various study design.
Tests for Association
Basic principle of GWAS
Depends on study design
• Case-control study
• Family-based study: case-parent triad, case-sib pairs being popular choices
• Longitudinal Cohort Study
• Looking at a secondary outcome under case-control sampling
The GWAS Mantra!
Primary Analysis
• Single marker association tests
• Genetic susceptibility model
- Dominant, recessive, co-dominant
• Which test to use
• Multiple testing correction
Case-Control Study: Standard Analysis
Pros and Cons
• Simple, Complete.
• Robust to misspecification of the true dominance pattern
• Less powerful.
• Unreliable for sparse table
Pros and Cons
• Test statistic has single df, so more powerful.
• Simple to report.
• Not robust to true mode of dominance
• Does not present entire information in the data.
Armitage’s trend test
• Test linear trend in log(OR) with # A allele
• Test statistic still has single d.f.
• Simplicity, use information from the 2 x 3 array
• More robust than 2 x 2 tests, but less robust than the 2 d.f. test.
Allelic test
• Previous tests were based on genotype
• Can also treat allele as the unit of observation.
• You have doubled the sample size!!
But…
• Serious impact on Type 1 error under departures from HWE
• Interpretation becomes trickier.
Example
AIC: Akaike information criterion, lower the value, better is model fit
Using logistic regression
• Trick: Just code genotype differently
• Dominant: G=1 if AA or Aa, 0 otherwise
• Recessive: G=1 if AA, 0 otherwise
• Trend: G=# A alleles, thus G=2 if AA, =1 if Aa and 0 if aa
• Two df test: Create two dummy variables:
G1=1 if Aa and 0 otherwise
G2=1 if AA and 0 otherwise
Perform likelihood ratio test of full (G1 and G2) vs reduced model (No G1, G2).
• Adjust for other variables, fit a multivariate model
Example
Flip Flops
• Under a co-dominant model you see a non-monotone trend, i.e.,
• OR(Aa)<1 and OR(AA)>1
• You will likely miss these under the trend test.
Alternative tests
• Use alternative maximal test statistic
• Calculate dominant, recessive, trend, codominabt: take maximum test statistic
• Use permutation to get right P-value
Caveats
• Resist temptations of going on to a fishing expedition. “MOST SIGNIFICANT CODING”
• Mode of inheritance models were developed for simple mendelian disease with near-complete penetrance, much more difficult to believe for complex diseases.
Reliable Test
• Co-dominant is model free
• Not much loss of power unless AA
(homozygous carriers) are very rare.
• Log additive is what is reported most of the time to risk some false positives but enhance power.
QQ Plots
• Nice visual tools for checking association and systematic biases
• Plot observed (-log10)P-values versus expected under the global null (i.e. quantiles draw from U[0,1])
• Since vast majority (>99%) of tested markers are not associated with the trait, plot should fall along y=x line (if we are lucky we will see a few departures in the tail).
• Departures could be due to stratification, cryptic relatedness, differential genotyping error, incorrect test.
Clear population stratification bias
Family Based Studies
• You heard about population stratification.
• Solutions:
-Match on self-reported ethnicity
-Adjust for Principal components extracted from markers.
-Use family based controls
Case-sib (conditional logistic regression)
Case-parent (TDT, FBAT)
Nuclear Families
Extended Pedigrees
Why use families
• Robust tests.
• Detect genotyping error with inheritance impossibilities. (mother and father AA, offspring Aa).
• Do not have to think about selection of
“good” controls.
Why not use families
• Case-control typically more powerful unless the disease is very rare.
• Not just logistic regression or trend chisquared as analysis tools.
• Much harder to recruit (depending on the disease : late onset or childhood disorder).
Hypotheses
• Case-control design
• Family-based designs have no power to detect association unless linkage is present.
• When testing for association in family-based design, HA is always: both linkage and association is present between the marker and disease susceptibility locus (DSL) underlying the trait.
The null hypotheses
Mendelian Transmission
Transmission Disequilibrium Test
• Spielman et al, AJHG, 1998
• H0: Association but no linkage
A simple statistical test
• No variation in diagonal elements
(homozygote parents have no uncertainty in determining the conditional genotype distribution of the offspring).
• Under the null (i.e., Mendelian transmission) x | x +y ~Binomial (n=x+y, p=1/2)
• Similarly, like McNemar’s test:
Test statistic:
TDT Example
FBAT : Family based association tests
• Extending TDT beyond case-parent trios
• Test statistic for FBAT mimics a natural covariance function between trait and genotype.
• i: family j: individual Sum over all i and j
• T: Trait (centered) X: Coded Genotype
• S: parental genotype or a sufficient statistic for parental genotype
Details
• The E(X|S) is calculated under Mendelian transmission.
• X-E(X|S): residual of the transmission of parental genotype to offspring.
• You basically assess whether there is any association between the trait and this genotype residual.
Test statistic
• Under all three nulls, E(U)=0.
Test Statistic:
• Note all expectation and variance are on X, conditional on parental genotype and trait T.
Another Tool: Conditional Likelihood
• Used in case-sib studies, in general in matched studies. Strata: Family/pair.
• Breslow et al (1978) first proposed this tool for matched case-control data.
• R function clogit does this. Underlying codes use survival model as there is connection with partial likelihood from Cox’s proportional hazard model.
• For case-sib studies, sib is the control and the genotype is the exposure. The contribution of a given pair to the conditional likelihood is:
• exp[ β Genotype(case)] exp[ β Genotype(case)]+exp[β Genotype(control)]
-Obtain variance-covariance of conditional MLE using inverse Fisher information.
Summary
• Different tests for association in casecontrol studies: 2 by 3 table and logistic regression.
• Family based studies: tests and hypotheses.
• Study design choices, power, recruitment.