Biotechnology - Cloudfront.net
advertisement

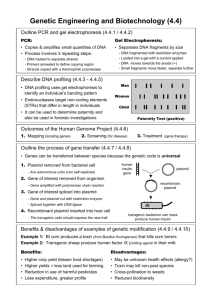
Biotechnology 3A1: DNA, and is some cases RNA, is the primary source of heritable information. Pair Share •What is biotechnology? •How long has biotechnology been taking place? •Is biotechnology and genetic engineering the same thing? •What does it mean to engineer something? Biotechnology vs Genetic engineering • Biotechnology: manipulation of organisms or their components to make useful products • Long history: selective breeding of farm animals; using microorganisms to make wine and cheese • Genetic Engineering: direct manipulation of genes for practical purposes • Launched a revolution in biotechnology expanding the scope of potential applications (agriculture, criminal law, medical research) • Examples: • • • • Recombinant DNA: DNA from two sources combined together Restriction Enzymes: essentially specific DNA cutters Gel electrophoresis: technique used to separate DNA by size Polymerase chain reaciton (PRC): technique used to quickly make copies of DNA Review – What is a gene???? • Chromosome = DNA • DNA = multiple genes • Gene = segments of DNA that codes for a protein • Protein = 3D structure of amino acids folded into a specific way that provides specific structure and function • To work directly with specific genes, scientists have developed techniques to clone DNA Gene Cloning • Gene Cloning: the produciton of multiple copies of a single gene; two basic purposes: to make copies of a particular gene and to produce a protein product • One common approach to gene cloning is to use bacteria • Review of bacterial DNA • large circular molecule of DNA • Plasmid: small circular DNA molecule that replicates separately from the bacterial chromosome – contains a small number of genes beneficial to the bacteria in particular environments, but not needed for survival and reproduction General Steps of Gene Cloning • Isolate plasmid and gene of interest • Insert gene of interest into plasmid • Return plasmid back into bacteria • Allow bacteria to reproduce reproduced bacteria will also replicate gene of interest • Products can be used to do further research of the particular gene, insert the gene into other organisms (recombinant DNA), use to clean up toxic waste, used for medical purposes Bacterium 1 Gene inserted into plasmid Bacterial chromosome Cell containing gene of interest Plasmid Recombinant DNA (plasmid) Gene of interest DNA of chromosome 2 Plasmid put into bacterial cell Recombinant bacterium 3 Host cell grown in culture to form a clone of cells containing the “cloned” gene of interest Gene of Interest Protein expressed by gene of interest Copies of gene Basic Protein harvested 4 Basic research and various applications research on gene Gene for pest resistance inserted into plants Gene used to alter bacteria for cleaning up toxic waste Protein dissolves blood clots in heart attack therapy Basic research on protein Human growth hormone treats stunted growth Restriction site Using Restriction Enzymes to Make Recombinant DNA • Restriction enzymes: Enzymes that cuts DNA at specific locations; also called restriction endonucleases • Recognizes specific DNA sequence called a restriction site • Cuts DNA strands at precise points within the restriction site • Most restriction sites are symmetrical, sequences of nucleotides is the same on both strands when read in the 5’3’ direction • Results in restriction fragments – pieces of DNA cut by restriction enzymes • All copies of a particular DNA molecule always yield the same set of restriction fragments when exposed to the same restriction enzyme • Most useful restriction enzymes cut in a staggered manner resulting in sticky ends • Sticky ends: short extensions of DNA that can form hydrogenbonded base pairs with complementary sticky ends on any other DNA molecule cut with the same restriction enzyme • DNA cut but the same restriction enzymes can be recombined to form recombinant DNA • DNA ligase: enzyme that catalyzes the formation of covalent bonds that close up the sugar-phosphate backbones of DNA; think back to Okazaki gragments during replication • https://www.youtube.com/watch?v=pDHCHa1C85Y DNA 1 5 3 3 5 Restriction enzyme cuts sugar-phosphate backbones. Sticky end 2 DNA fragment added from another molecule cut by same enzyme. Base pairing occurs. One possible combination 3 DNA ligase seals strands. Recombinant DNA molecule Cloning a Eukaryotic Gene in a Bacterial Plasmid (More Details) • Cloning Vector: DNA molecule that can carry foreign DNA into a host cell and replicate there • Bacteria plasmids are widely used as cloning vectors because they are easily isolated from bacteria, manipulated to form recombinant plasmids by inserting foreign DNA, and reintroduced into bacterial cells; they also multiple rapidly • Due to the difference in the way genes are expressed in eukaryotic cells vs prokaryotic cells, it is difficult to get a eukaryotic gene to be expressed in bacteria. • Alternative is to use yeast, which is a eukaryotic cell, but also contains a plasmid Cloning a Eukaryotic Gene in a Bacterial Plasmid • In gene cloning, the original plasmid is called a cloning vector • A cloning vector is a DNA molecule that can carry foreign DNA into a host cell and replicate there Producing Clones of Cells Carrying Recombinant Plasmids • Several steps are required to clone the hummingbird β-globin gene in a bacterial plasmid: • The hummingbird genomic DNA and a bacterial plasmid are isolated • Both are digested with the same restriction enzyme • The fragments are mixed, and DNA ligase is added to bond the fragment sticky ends Animation: Cloning a Gene • Some recombinant plasmids now contain hummingbird DNA • The DNA mixture is added to bacteria that have been genetically engineered to accept it • The bacteria are plated on a type of agar that selects for the bacteria with recombinant plasmids • This results in the cloning of many hummingbird DNA fragments, including the β-globin gene Fig. 20-4-1 Hummingbird cell TECHNIQUE Bacterial cell lacZ gene Restriction site ampR gene Bacterial plasmid Sticky ends Gene of interest Hummingbird DNA fragments Fig. 20-4-2 Hummingbird cell TECHNIQUE Bacterial cell lacZ gene Restriction site ampR gene Sticky ends Bacterial plasmid Gene of interest Hummingbird DNA fragments Nonrecombinant plasmid Recombinant plasmids Fig. 20-4-3 Hummingbird cell TECHNIQUE Bacterial cell lacZ gene Restriction site ampR gene Sticky ends Bacterial plasmid Gene of interest Hummingbird DNA fragments Nonrecombinant plasmid Recombinant plasmids Bacteria carrying plasmids Fig. 20-4-4 Hummingbird cell TECHNIQUE Bacterial cell lacZ gene Restriction site ampR gene Sticky ends Bacterial plasmid Gene of interest Hummingbird DNA fragments Nonrecombinant plasmid Recombinant plasmids Bacteria carrying plasmids RESULTS Colony carrying nonrecombinant plasmid with intact lacZ gene Colony carrying recombinant plasmid with disrupted lacZ gene One of many bacterial clones Screening a Library for Clones Carrying a Gene of Interest • A clone carrying the gene of interest can be identified with a nucleic acid probe having a sequence complementary to the gene • This process is called nucleic acid hybridization • A probe can be synthesized that is complementary to the gene of interest • For example, if the desired gene is 5 … G G C T A A C T T A G C … 3 – Then we would synthesize this probe 3 C C G A T T G A A T C G 5 • The DNA probe can be used to screen a large number of clones simultaneously for the gene of interest • Once identified, the clone carrying the gene of interest can be cultured Fig. 20-7 TECHNIQUE Radioactively labeled probe molecules Multiwell plates holding library clones Probe DNA Gene of interest Single-stranded DNA from cell Film • Nylon membrane Nylon Location of membrane DNA with the complementary sequence Amplifying DNA in Vitro: The Polymerase Chain Reaction (PCR) • The polymerase chain reaction, PCR, can produce many copies of a specific target segment of DNA • A three-step cycle—heating, cooling, and replication—brings about a chain reaction that produces an exponentially growing population of identical DNA molecules Fig. 20-8 5 TECHNIQUE 3 Target sequence 3 Genomic DNA 1 Denaturation 5 5 3 3 5 2 Annealing Cycle 1 yields 2 molecules Primers 3 Extension New nucleotides Cycle 2 yields 4 molecules Cycle 3 yields 8 molecules; 2 molecules (in white boxes) match target sequence Fig. 20-8a 5 TECHNIQUE 3 Target sequence Genomic DNA 3 5 Fig. 20-8b 1 Denaturation 5 3 3 5 2 Annealing Cycle 1 yields 2 molecules Primers 3 Extension New nucleotides Fig. 20-8c Cycle 2 yields 4 molecules Fig. 20-8d Cycle 3 yields 8 molecules; 2 molecules (in white boxes) match target sequence Concept 20.2: DNA technology allows us to study the sequence, expression, and function of a gene • DNA cloning allows researchers to • Compare genes and alleles between individuals • Locate gene expression in a body • Determine the role of a gene in an organism • Several techniques are used to analyze the DNA of genes Gel Electrophoresis and Southern Blotting • One indirect method of rapidly analyzing and comparing genomes is gel electrophoresis • This technique uses a gel as a molecular sieve to separate nucleic acids or proteins by size • A current is applied that causes charged molecules to move through the gel • Molecules are sorted into “bands” by their size Video: Biotechnology Lab Fig. 20-9 TECHNIQUE Mixture of DNA molecules of different sizes Power source – Cathode Anode + Gel 1 Power source – + Longer molecules 2 RESULTS Shorter molecules Fig. 20-9a TECHNIQUE Mixture of DNA molecules of different sizes Power source Anode – Cathode + Gel 1 Power source – + Longer molecules 2 Shorter molecules Fig. 20-9b RESULTS • In restriction fragment analysis, DNA fragments produced by restriction enzyme digestion of a DNA molecule are sorted by gel electrophoresis • Restriction fragment analysis is useful for comparing two different DNA molecules, such as two alleles for a gene • The procedure is also used to prepare pure samples of individual fragments Fig. 20-10 Normal -globin allele 175 bp DdeI Sickle-cell allele Large fragment 201 bp DdeI Normal allele DdeI DdeI Large fragment Sickle-cell mutant -globin allele 376 bp DdeI 201 bp 175 bp Large fragment 376 bp DdeI (a) DdeI restriction sites in normal and sickle-cell alleles of -globin gene DdeI (b) Electrophoresis of restriction fragments from normal and sickle-cell alleles