Supplementary Materials anf Methods (doc 37K)
advertisement

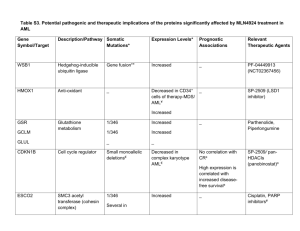
Supplementary Information - Materials and Methods AML patient and healthy donor materials Peripheral blood mononuclear cells (PBMC) or bone marrow mononuclear cells (BMMNC) were collected from healthy donors or AML patients enrolled in the UK MRC/NCRI AML 14, 15, 16 and 17 clinical trials at point of diagnosis, before treatment and following informed consent from patients in accordance with the 1964 Declaration of Helsinki (refer to Supplemental Table S1). Mononuclear cells used in this study were separated on Ficoll-Hypaque (Sigma, Poole Dorset U.K) as previously described.1 Cell surface immunophenotyping of AML patient blasts and Tregs PBMC/BMMNC from AML patients or healthy donors were thawed and washed in pre-warmed T-cell medium consisting of Iscove’s modified Dulbecco’s Medium supplemented with 10% heat inactivated human AB-serum (Invitrogen, Paisley, U.K.). Cells (106) were washed in staining buffer (PBS, 2% FCS, 1mM EDTA, 0.02% NaN3) before incubation with the following anti-human antibodies purchased from BD Bioscience U.K. unless otherwise stated; CD45-FITC, CD200-PE, CD34-PerCP-Cy5.5 for AML blast-cell phenotyping. For Treg cell analysis, PBMC or BMMNC were incubated in staining buffer in the presence of CD4-PerCP-Cy5.5, CD3-APC and CD25-PE (MiltenyiBiotec, Surrey, UK) followed by a fix and permeabilization step using Foxp3 Fixation/Permeabilization Buffer according to the manufacturer’s instruction (eBiosciences, Hatfield U.K.). Subsequently, cells were incubated with FoxP3-FITC (clone PCH101) in the presence of 2% normal rat serum. All incubations were carried out in 100µl of staining buffer at 4ºC for 30min in the dark with the exception of FoxP3 which was carried out in 50µl for 20min. Cells were analyzed by flow cytometry (see data acquisition and analysis). Isolation of Treg cells from healthy donors or AML patients Treg cells were isolated from PBMC/BMMNC of healthy donors or AML patients using a CD4+CD25+Treg cell isolation kit in accordance with manufacturer’s instruction (MiltenyiBiotec). Briefly, cryo-preserved cells were thawed and subjected to gradual resuspension in 10ml processing buffer (PBS, 5 mM MgCl2, adjusted to contain 1 mg/ml DNase) at a rate of 1 ml/min. Following two washes with chilled column buffer (PBS, 1% BSA, 2 mM EDTA), a biotin antibody cocktail mix containing anti-human CD8, CD14, CD16, CD19, CD36, CD56, CD123, TCRγ/δ and CD235a was added to 107 cells and incubated for 10 min at 4oC. Afterwards, anti-biotin micro beads were added and incubated for 15 min, prior to sequential washes with 1 ml column buffer. The cells were separated on an LD column (MiltenyiBiotec) and the effluent incubated with anti-human CD25 microbeads for 15 min at 4oC. Cells were washed with column buffer and separated on an MS column (MiltenyiBiotec) into CD4+CD25- and CD4+CD25+ fractions. Enrichment of Treg cells was verified by immunophenotyping (see cell surface immunophenotyping of AML patient blasts and Treg cell subpopulations). T-cell proliferation assay using thymidine incorporation Isolated CD4+CD25++Treg effector cells from AML patients or healthy donors were incubated with CD4+CD25- responder cells at the following responder to Treg effector ratios (1 to 1, 1 to 0.1, 1 to 0.01, 1 to 0.001 and 1 to 0.0001) for 54 h at 37ºC/5% CO2 in T-cell medium containing 1 μg/ml anti-human CD28 (BD, clone 28.2) in anti-CD3 (eBiosciences, clone OKT3) coated 96 well plates. Following incubation, 0.034 MBq [methyl-3H]-thymidine (Amersham, UK) in 50 μl pre-warmed T-cell medium was added to each well before a further 18 h incubation. Cells were subsequently harvested onto printed Filtermat-A filters and Meltex-A melt-on scintillation sheets prior to analysis using a Trilux-1450 scintillation counter (Perkin-Elmer, Cambridge U.K.). Data acquisition and analysis Data were acquired using an Accuri C6 cytometer (Accuri, St Ives, U.K.) or a FACSCanto II cytometer (BD). Data analysis was performed using FCS express v4.0 (DeNovo Software, Canada). Background fluorescence was established by isotype-matched controls. For analysis of CD200 expression on AML patient blasts, lymphocytes were excluded from the analysis based on low side scatter (SSC) and high CD45 expression. CD200 expression level relates to CD200lo data inclusive of minimum to Q1 (0-0.68) and CD200hi data inclusive of Q3 to maximum (2.87-16.14) as previously described.2 For analysis of Treg cells a combination of CD3, CD4 and CD25 was used to define this population which were CD3+CD4+CD25++ (>95% FoxP3+). For intracellular cytokine analysis AquaLive was used to discriminate dead cells. The presence of TNFα, IL2 and IFNγ was assessed in the CD4+ T-cell pool. Significance of difference was determined using oneway ANOVA with Tukey’s post-hoc multiple testing correction. Linear regression using Pearson’s correlation coefficient was performed to establish data correlations. All statistical analyses were performed using Sigma plot v10 (Stystat Software, CA, U.S.A.) Supplementary Information references 1 Tonks A, Tonks AJ, Pearn L, Pearce L, Hoy T, Couzens S, et al. Expression of AML1ETO in human myelomonocytic cells selectively inhibits granulocytic differentiation and promotes their self-renewal. Leukemia 2004; 18: 1238-45. 2 Coles SJ, Wang EC, Man S, Hills RK, Burnett AK, Tonks A, et al. CD200 expression suppresses natural killer cell function and directly inhibits patient anti-tumor response in acute myeloid leukemia. Leukemia 2011; 25: 792-9.