BIO240 Exam #3 SP13 answer key
advertisement

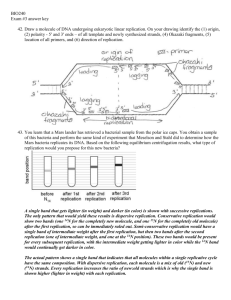
Exam #3 Answer key 44. Consider the DNA sequence 3′ TACCGTGCGTGACATTAAGCC 5′ a. What would be the sequence of the complementary strand of DNA? 5ʹATGGCACGCACTGTAATTCGG 3ʹ b. What would be the sequence of an RNA produced by using the DNA sequence shown as the template? 5ʹAUGGCACGCACUGUAAUUCGG 3ʹ c. Some viruses contain an enzyme that replicates RNA from RNA. Using the RNA sequence from part b. as the template, what would be the sequence of the complementary RNA? 3′ UACCGUGCGUGACAUUAAGCC 5′ 45. Describe nucleotide structure. Contrast differences between RNA and DNA nucleotides, include specific components and their attachment sites to one another. A nucleotide is composed of three components – a pentose (five carbon sugar), a phosphate group, and a nitrogenous base. The pentose sugar in RNA is ribose, and is deoxyribose in DNA. The difference is at the 2’ carbon. (‘ indicating carbon in the sugar) – ribose has an –OH group while deoxyribose has a –H. The negatively charged phosphate group (PO4) is covalently bonded at the 5’ carbon. Nitrogenous bases are classified as either purines (double-ringed) or pyrimidines (single rings). Adenine and guanine are purines while cytosine, thymine, and uracil are pyrimidines. Thymine is present only in DNA while uracil is only present in RNA. The nitrogenous base is covalently bonded to the 1’ carbon. NOTE: Many of you wanted to explain about complementary base pairing, how nucleotides are bonded together, etc, BUT this is NOT what the question is asking. 46. You learn that a Mars lander has retrieved a bacterial sample from the polar ice caps. You obtain a sample of this bacteria and perform the same kind of experiment that Meselson and Stahl did to determine how the Mars bacteria replicates its DNA. Based on the following equilibrium centrifugation results, what type of replication would you propose for this new bacteria? A single band that gets lighter (in weight) and darker (in color) is shown with successive replications. The only pattern that would yield these results is dispersive replication. Conservative replication would show two bands (one 14N for the completely new molecule, and one 15N for the completely old molecule) after the first replication, so can be immediately ruled out. Semi-conservative replication would have a single band of intermediate weight after the first replication, but then two bands after the second replication (one of intermediate weight, and one at the 14N position). These two bands would be present for every subsequent replication, with the intermediate weight getting lighter in color while the 14N band would continually get darker in color. The actual pattern shows a single band that indicates that all molecules within a single replicative cycle have the same composition. With dispersive replication, each molecule is a mix of old (15N) and new (14N) strands. Every replication increases the ratio of new:old strands which is why the single band is shown higher (lighter in weight) with each replication. 47. Draw a molecule of DNA undergoing eukaryotic linear replication. On your drawing identify the (1) origin, (2) polarity - 5′ and 3′ ends – of all template and newly synthesized strands, (4) Okazaki fragments, (5) location of all primers, and (6) direction of replication. Note: replication bubble contains the origin of replication in the MIDDLE (on half being a replication fork). Which strand is the leading strand or the lagging strand will change on the other replication fork. 48. Below is a linear sequence of DNA that you want to amplify. (spaces are shown to make reading sequences easier) 5′ AGGCGGTTAG GCCATGCTTA CCGGCATTGG CCGGCCTAAA GGCCAATTTT 3′ 3′ TCCGCCAATC CGGTACGAAT GGAAGTAACC GGCCGGATTT CCGGTTAAAA 5′ (as mentioned in class, these two As should be two Cs … but did not make a difference with the results) After you perform PCR, you digest the fragment with the enzyme HaeIII. You then separate the fragments by running them on a 4% agarose gel. You use a marker with known fragment sizes of 5bp, 10bp, 15bp, 20bp, 30bp, 40bp, and 50bp. The enzyme Hae III has the recognition site 5′ GGˇCC 3′ 3′ CCˆGG 5′ a. Design the 10 nucleotide-length primers needed to amplify this region – provide the sequence and polarity (5ʹ and 3ʹ ends) Primers provide the beginning of a new strand, and must be read in the 5’ to 3’ direction since nucleotides must be added to the 3’ end. This determines how the primers are designed. Primer sequences are shown in blue. 5′ AGGCGGTTAG GCCATGCTTA CCGGCATTGG CCGGCCTAAA GGCCAATTTT 3′ ←. . . 3’ CCGGTTAAAA 5’ 5’ AGGCGGTTAG 3’ … (elongation continues ….) → 3′ TCCGCCAATC CGGTACGAAT GGCCGTAACC GGCCGGATTT CCGGTTAAAA 5′ b. Indicate the restriction sites on the fragment. HaeIII has a recognition site of 5’ GG/CC 3’ 3’ CC/GG 5’ This enzyme creates blunt ends, as the cut is at the same location on both strands. The restriction site is highlighted in red. 5′ AGGCGGTTAG G/CCATGCTTA CCGGCATTGG/ CCGG/CCTAAA GG/CCAATTTT 3′ 3′ TCCGCCAATC C/GGTACGAAT GGCCGTAACC/ GGCC/GGATTT CC/GGTTAAAA 5′ Fragment Size 11bp 19bp 4bp 8bp 8bp Since there are 4 restriction sites, digestion yields 5 fragments. However, since two fragments are the same size, they will migrate together on a gel. The 8bp fragment would be expected to be darker and thicker than the others, since it contains twice as much DNA. c. Draw the gel as it would appear with the marker in the first lane, the uncut fragment in the second lane, and the cut fragment in the third lane. Wells ▬ ▬ 50bp ▬ ▬ 40bp ▬ 30bp ▬ 20bp 15bp 10bp 5bp ▬ ▬ ▬ ▬ ▬ ▬ ▬ ▬ ▬ 49. Explain the 3 necessary components of a cloning vector. Cloning vectors are small, circular DNA molecules that are used to make multiple copies of a gene of interest. The following components are required: #1 Origin of replication – a AT rich region that serves as the starting site for DNA replication (a cloning vector needs to replicate independently) #2 Unique restriction site – a cloning vector needs to contain ONE recognition site for a particular enzyme. Digestion with enzyme will open up the vector; digestion of gene of interest with the same endonuclease will yield sticky ends with identical sequences. Annealing gene with vector will cause some copies to insert into vector. #3 Selectable marker – this is often antibiotic resistance, which serves as an easy way to differentiate cells that have taken up the vector from non-recombinant cells. 50. Compare and contrast the two basic types of terminators found in bacterial cells in regard to both structure and function. The two terminator types are Rho-independent and Rho-dependent. Rho-independent – transcribed RNA contains inverted repeats, which form a hairpin secondary structure. This is followed by a polyA region on the template DNA which will transcribe into a polyU region. The presence of the hairpin slows the speed of RNA polymerase. This reduction in speed, coupled by the weak (2 hydrogen bonds between each A-U base pair) bonds, causes RNA to peel away from its template, and transcription halts. Rho-dependent – transcribed RNA likewise contains inverted repeats, which form a hairpin secondary structure. The following sequence is irrelevant. Rho is an enzyme with helicase activity. Rho protein binds to unstructured (linear) RNA molecule, and travels in 5’ to 3’ direction. The hairpin will slow down RNA polymerase. This allows Rho to “catch up” to where RNA is still hydrogen bonded to its template DNA strand. Rho breaks the hydrogen bonds (regardless of number), causing RNA to be released from its template.