PGLO - jvbiologyk
advertisement

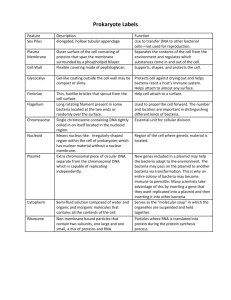
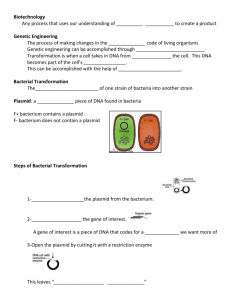
pGLO Transformation PreLab Introduction to Transformation In this lab you will perform a procedure known as a genetic transformation. Remember that a gene is a piece of DNA which provides the instructions for making (coding for) a protein which gives an organism a particular trait. Genetic transformation literally means change caused by genes and it involves the insertion of gene(s) into an organism in order to change the organism's trait(s). Genetic transformation is used in many areas of biotechnology. In agriculture, genes coding for traits such as frost, pest, or spoilage resistance can be genetically transformed into plants. In bioremediation, bacteria can be genetically transformed with genes enabling them to digest oil spills. In medicine, diseases caused by defective genes are beginning to be treated by gene therapy; that is, by genetically transforming a sick person's cells with healthy copies of the gene involved in their disease. You will use a procedure to transform bacteria with a gene that codes for a Green Fluorescent Protein (GFP). The real-life source of this gene is the bioluminescent jellyfish Aequorea victoria. The gene codes for a Green Fluorescent Protein which causes the jellyfish to fluoresce and glow in the dark. Following the transformation procedure, the bacteria will express their newly acquired jellyfish gene and produce the fluorescent protein which will cause them to glow a brilliant green color under ultraviolet light. In this activity, you will learn about the process of moving genes from one organism to another with the aid of a plasmid. In addition to one large chromosome, bacteria naturally contain one or more small circular pieces of DNA called plasmids. Plasmid DNA usually contains genes for one or more traits that may be beneficial to bacterial survival. In nature, bacteria can transfer plasmids back and forth allowing them to share these beneficial genes. This natural mechanism allows bacteria to adapt to new environments. The recent occurrence of bacterial resistance to antibiotics is due to the transmission of plasmids. Bio-Rad's unique pGLO plasmid encodes the gene for the Green Fluorescent Protein (GFP) and a gene for resistance to the antibiotic; ampicillin. pGLO also incorporates a special gene regulation system which can be used to control expression of the fluorescent protein in transformed cells by adding the sugar, arabinose, to the cells' nutrient medium. Selection for cells that have been transformed with pGLO DNA is accomplished by growth on antibiotic plates. Transformed cells will appear white (wild type phenotype) on plates not containing arabinose, and fluorescent green when arabinose is included in the nutrient agar. Your task will be: 1. To do the genetic transformation. 2. To determine the degree of success in your efforts to genetically alter an organism Pre-LAB Questions #1-5 There are many considerations that need to be thought through in the process of planning a scientific laboratory investigation. Below are a few for you to ponder as you take on the challenge of doing a genetic transformation. Since scientific laboratory investigations are designed to get information about a question, our first step might be to formulate a question for this investigation. Can I Genetically Transform an Organism? Which Organism? 1. To genetically transform an entire organism, you must insert the new gene(s) into every cell in the organism. Which organism is better suited for total genetic transformation - one composed of many cells, or one composed of a single cell? 2. Scientists often want to know if the genetically transformed organism can pass its new traits on to its offspring and future generations. To get this information, which would be a better candidate for your investigation, an organism in which each new generation develops and reproduces quickly, or one which does this more slowly? 3. Safety is another important consideration in choosing an experimental organism. What traits or characteristics should the organism have (or not have) to be sure it won't harm you or the environment? 4. Based on the above considerations, which would be the best choice for a genetic transformation: bacteria, earthworm, fish, or mouse? 5. Explain your reasoning for the above answer (#4). How Can I Tell if Cells Have Been Genetically Transformed? Recall that the goal of genetic transformation is to change an organism's traits (phenotype). However, before any change in the phenotype of an organism can be detected, a thorough examination of its natural (pre-transformation) phenotype must be made. 6. Look at the colonies of E.coli on your starter plates. The following pre-transformation observations of E. coli might provide a basis, to make reference to, when attempting to determine if any genetic transformation has occurred . List all observable traits or characteristics that can be described in the non-transformed cells: (# of colonies, size of the largest/smallest colony, color of the colonies, distribution/sketch of the colonies on the plate, visible appearance when viewed with ultraviolet (UV) light, ability of the cells to live and reproduce in the presence of an antibiotic such as ampicillin, etc) 7. Describe how you could use two LB agar plates, some E. coli and some ampicillin to determine how E. coli cells are affected by ampicillin. 8. What would you expect your experimental results to indicate about the effect ampicillin has on the E. coli cells? The Genes Genetic transformation involves the insertion of some new DNA into the E. coli cells. In addition to one large chromosome, bacteria often contain one or more small circular pieces of DNA called plasmids. Plasmid DNA usually contains genes for more than one trait. Scientists can use a process called genetic engineering to insert genes coding for new traits into a plasmid. In this case, the pGLO plasmid carries the gene (GFP) which produces the green fluorescent protein and a gene (bla) that codes for a protein giving bacteria resistance to an antibiotic. The genetically engineered plasmid can then be used to genetically transform bacteria to give then this new trait(s). The Act of Transformation This transformation procedure involves three main steps. These steps are intended to introduce the plasmid DNA into the E. coli cells and provide an environment for the cells to express their newly acquired genes. To move the plasmid DNA - pGLO through the cell membrane you will: 1. Use a transformation solution of CaCl2 (calcium chloride) Opens nuclear membrane thus 2. Carry out a procedure referred to as "heat shock" exposing DNA plasmid For transformed cells to grow in the presence of ampicillin you must: 3. Provide them with nutrients and a short incubation period to begin expressing their newly acquired genes. Transformation Lab Your workstation should have the following material and supplies: *E coli starter plates *1 LB agar plate *1 LB/amp/ara agar plate *Transformation solution *7 inoculation loops *pipettes *cup of ice *marking pen *2 LB/amp agar plates *LB broth *1 foam microtube holder *42oC water bath In addition, there is a common station with the following supplies: * 1 vial Hydrated pGLO plasmid *37oC incubator Transformation Procedure- be sure to use sterile techniques! 1. Label closed micro test tube +DNA & another -DNA. Label tubes with your group's name. Place them in the foam tube holder. 2. Open the tubes and using a sterile transfer pipette, transfer 250 l of Transformation Solution (CaCl2) into each tube. 3. Place the tubes on ice. 4. Use a sterile loop to pick up one single colony of bacteria from your starter plate. Pick up the +DNA tube and immerse the loop into the Transformation Solution at the bottom of the tube. Spin the loop between your index finger and thumb until the entire colony is dispersed in the Transformation Solution (no floating chunks). Place the tube back in the tube rack in the ice. Use a new sterile loop, repeat the procedure for the -DNA tube. 5. Examine the pGLO DNA solution with the UV lamp. Note your observations. Immerse a new sterile loop into the plasmid DNA stock tube. Withdraw a loop full. There should be a film of plasmid solution across the ring. This is similar to seeing a soapy film across a ring for blowing soap bubbles. Mix the loop full into the cell suspension of the +DNA tube. Close the tube. Close the tube and return it to the rack on ice. Also close the -DNA tube. Do not add plasmid DNA to the -DNA tube. Why not? 6. Incubate the tubes on ice for 10 minutes. Make sure to push the tubes all the way down in the rack so the bottom of the tubes sick out and make contact with the ice. 7. While the tubes are sitting on ice, label your four agar plates on the bottom (not the lid) as follows: *Label one LB/amp plate: +DNA *Label the LB/amp/ara plate: +DNA *Label the other LB/amp plate: -DNA *Label the LB plate: -DNA 8. Heat shock. Using the foam rack as a holder, transfer both the (+) and (-) tubes into the 42oC water bath for exactly 50 seconds. Make sure to push the tubes all the way down in the rack so the bottom of the tubes stick out and make contact with the warm water. When the 50 seconds are done, place both tubes back on ice. For the best transformation results, the change for the ice (0oC) to 42oC and then back to the ice must be rapid. Incubate tubes on ice for 2 minutes. 9. Remove the rack containing the tubes from the ice and place on the desk top. Open a tube and, using a new sterile pipette, add 250 l of LB broth to the tube and re-close it. Repeat with a new sterile pipette for the other tube. Incubate the tubes for 10 minutes at room temperature. 10. Tap the closed tubes with your finger to mix. Using a new sterile pipette for each tube, pipette 100 ml of the transformation and control suspensions onto the appropriate plates. 11. Use a new sterile loop for each plate. Spread the suspensions evenly around the surface of the agar by quickly skating the flat surface of a new sterile loop back and forth across the plate surface. 12. Stack your plates and tape then together. Put your group name and class period on the bottom of the stack and place it upside down in the 37oC incubator for 24 hours. Review Questions After completing the lab but before collecting data and analyzing your results, answer the following questions: 9. On which of the plates would you expect to find bacteria most like the original non transformed E. coli colonies you initially observed? 10. Explain your predictions: 11. If there are any genetically transformed bacterial cells, on which plate(s) would they most likely be located? 12. Explain your predictions: 13. Which plates should be compared to determine if any genetic transformation has occurred? 14. Why? 15. What is meant by "control plate"? Which plate would be the "control plate"? 16. What purpose does a control serve? Data Collection and Analysis Observe the results you obtained from the transformation lab under normal room lighting. Then turn out the lights and hold the ultraviolet light over the plates. Observe and draw what you see on each of the four plates carefully. Put your drawings in the data table in the column on the right. Record your data to allow you to compare observations of the "+DNA" cells with those you record for the non-transformed E. coli. Write down the following observations for each plate. OBSERVATIONS (# of colonies, color, amount of growth, etc.) +DNA LB/amp +DNA LB/amp/ara OBSERVATIONS (# of colonies, color, amount of growth, etc.) -DNA LB/amp -DNA LB The goal of data analysis for this investigation is to determine if the data indicate that genetic transformation has occurred. 17. Which of the traits that you originally observed for E.coli did not seem to become altered? In the space below list these non-transformed traits and how you arrived at this analysis for each trait. Original trait Analysis of observations 18. Of the E.coli traits you originally noted, which seem now to be significantly different after performing the transformation procedure? List those traits below and describe the changes that you observed. New Trait Observed Change 19. If the genetically transformed cells have acquired the ability to live in the presence of the antibiotic ampicillin, then what might be inferred about the other genes on the plasmid that you used in your transformation procedure? 20. From the results that you obtained, how could you prove that these changes that occurred were due to the procedure that you performed? What's Glowing? 21. If a fluorescent green color is observed in the E.coli colonies then a new question might well be raised, "What are the two possible sources of fluorescence within the colonies when exposed to UV light?" 22. Explain each: 23. Recall what you observed when you shined the UV-light source onto a sample of original plasmid DNA prior to completing the lab. Describe your observations. 24. Which of the two possible sources of the fluorescence can now be eliminated? 25. What does this observation indicate about the source of the fluorescence? 26. Describe the evidence that indicates whether your attempt at performing a genetic transformation was successful or not successful. The Interaction Between Genes and Environment 27. Look again at your four plates. Do you observe E.coli bacteria growing on a plain LB plate? 28. From your results, can you tell if these bacteria are ampicillin resistant by looking at them? 29. Explain your answer: 30. How would you change the bacteria's environment - the plate they are growing on - to best tell if they are ampicillin resistant? Very often an organism's traits are caused by a combination of its genes and its environment. Think about the green color you saw in the genetically transformed bacteria: 31. What two factors must be present in the bacteria's environment for you to see the green color? (Hint: one factor is in the plate and other factor is in how you look at the bacteria.) 32. What do you think each of the two environmental factors you listed above are doing to cause the genetically transformed bacteria to turn green? 33. What advantage would there be for an organism to be able to turn on or off particular genes in response to certain conditions? 34. The following is a segment of DNA that contains, within it, the gene for production of human insulin. Below that is a plasmid from the E. coli bacteria. Below that is a series of restriction enzymes and their recognition/cut sites: Your task is to determine which enzyme(s) could be used on both the insulin sequence and the plasmid to obtain the desired "sticky ends" that will then allow the gene to be successfully spliced into the plasmid. Explain your answer thoroughly and draw any explanation on the plasmid map above. 35. Below is a restriction map for the plasmid pGEN101 (total length = 20,000 bp). Using this map as a guide, give the number of restriction fragments along with their associated lengths that would result from digesting pGEN101 with the restriction enzymes EcoRI, BamHI, and a combination of EcoRI + BamHI 36. Two freshman Biology K students, interested in becoming gene jocks, performed the following set of restriction digests on a newly discovered plasmid pBLA230. The reactions they carried out, along with the fragments obtained in single double digests reactions were: Using this information, construct a restriction map of pBLA230. 37. sizes Lane 1 Lane 2 Lane 3 Plasmid pBR607 is a 26,000 plasmid containing ampicillin and tetracycline resistance genes, and restriction sites for the restriction enzymes EcoRI, BamHI, and PstI. Given the restriction map below for pBR607, show where the approximate positions of the restriction fragments (created from the following restriction digests: (A) EcoRI + PstI, (B) EcoRI + BamHI, and (C) EcoRI + BamHI + PstI) would appear - relative to each other - on a gel. Draw a gel showing the fragments created and the approximate size of each fragment following gel electrophoresis. Put (A) in lane 1, (B) in lane 2, and (C) in lane 3.