Supplementary Data 2 (docx 193K)

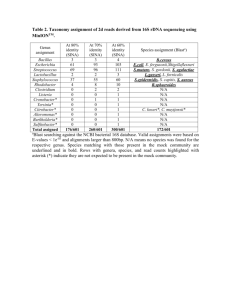
Two of the ribosomal sequence from the bacterial isolates matched the representative sequence of 2 of 3 dominant OTUs assembled from the 454 pyrosequencing data. DNA from the isolates was isolated and amplified using the standard 27F -1492R primer pair.
PCR conditions were as follows 5 minutes 94 ° C, 30 cycles of 94 ° C for 30 sec, 57 ° C for 45 sec and 72 ° C for 90 sec with the final elongation step at 72 ° C for 7 minutes. The PCR product was purified and sequenced using Sanger technology. Achromobacter isolated from the M. truncatula rhizosphere, when compared using BLAST, showed 99 % similarity against a representative sequence from OTU 19 (score of 544, e value = 1e-
159). Arthrobacter sp. isolated from the B. distachyon rhizosphere, when compared using BLAST, showed 100 % similarity against a representative sequence from OTU 39
(score of 556, e value = 7e-163). Alignments were based on the V1-V2 16S rRNA gene only. Genomic DNA was isolated from both strains and they were sequenced using half a plate of 454Flx each. Achromobacter gDNA sequencing produced 50 contigs (of which
14 were smaller than 100 bp in length). Arthrobacter gDNA sequencing produced 48 contigs (of which 17 were smaller than 100 bp in length).
The bacterium isolated from the rhizosphere of Medicago was identified as
Achromobacter xylosoxidans (top hit in the NCBI database – accession number:
NR_074754.1, based on the full 16S rRNA sequence and the colony isolated from the
Brachypodium rhizosphere was identified as Arthrobacter sp. (top hit in the NCBI database – accession number EF110914.1, based on the full 16S rRNA sequence – gDNA sequencing (Figure 1). Full 16S rRNA gene sequences were obtained from the genomic
DNA FASTA file using online RNAmmer 1.2 server (a part of CBS prediction server)
(Lagesen et al 2007). However, using the full 16S rRNA gene sequences would skew the results towards fully sequenced strains of these species, so in order to fully understand the phylogeny of these strains a partial 16S rRNA gene (1460 and 1486 bp of sequence starting at 27 bp from the start of the gene for Achromobacter and Arthrobacter, respectively) was compared (BLAST) against the GenBank database.
A
Achromobacter xylosoxidans
Achromobacter xylosoxidans subsp. xylosoxidans strain A19 16S ribosomal RNA
isolate 16S rRNA gene
Achromobacter xylosoxidans A8 complete genome
Achromobacter xylosoxidans A8 complete genome
Achromobacter xylosoxidans A8 strain A8 16S ribosomal RNA
Achromobacter sp. F32 16S
Achromobacter sp. R-46660
Arsenite-oxidizing bacterium Alcaligenes fecalis (HLE)
Alcaligenes sp. 16S rRNA
Achromobacter spp.
Alcaligenes faecalis strain N05
Achromobacter spp.
Achromobacter spp.
Achromobacter spp.
Achromobacter spp.
Achromobacter spp.
Bordetella
Pigmentiphaga daeguensis strain ML-3
Pusillimonas sp. YC6271
Rhodobacter
Ochrobactrum pseudogrignonense strain 21-6PIN
Tetrathiobacter kashmirensis strain 3T5F
Denitrobacter sp. CHNCT17
Burkholderia sp.
Kerstersia gyiorum strain HF2
Burkholderiales
Limnobacter thiooxidans strain CS-K2
Cupriavidus sp. ASC-9842
Pseudomonas lemoignei strain ATCC 17989T
Undibacterium sp. CMJ-15
Oxalobacteraceae
Achromobacter isolate 16S rRNA sequence used for comparison:
ACGCTAGCGGGATGCCTTACACATGCAAGTCGAACGGCAGCACGGACTTCGGTCTGGTGGCGAGTGGCGAACGGGTGAGTAAT
GTATCGGAACGTGCCTAGTAGCGGGGGATAACTACGCGAAAGCGTAGCTAATACCGCATACGCCCTACGGGGGAAAGCAGGGG
ATCGCAAGACCTTGCACTATTAGAGCGGCCGATATCGGATTAGCTAGTTGGTGGGGTAACGGCTCACCAAGGCGACGATCCGT
AGCTGGTTTGAGAGGACGACCAGCCACACTGGGACTGAGACACGGCCCAGACTCCTACGGGAGGCAGCAGTGGGGAATTTTGG
ACAATGGGGGAAACCCTGATCCAGCCATCCCGCGTGTGCGATGAAGGCCTTCGGGTTGTAAAGCACTTTTGGCAGGAAAGAAA
CGTCATGGGCTAATACCCCGTGAAACTGACGGTACCTGCAGAATAAGCACCGGCTAACTACGTGCCAGCAGCCGCGGTAATACG
TAGGGTGCAAGCGTTAATCGGAATTACTGGGCGTAAAGCGTGCGCAGGCGGTTCGGAAAGAAAGATGTGAAATCCCAGAGCTC
AACTTTGGAACTGCATTTTTAACTACCGGGCTAGAGTGTGTCAGAGGGAGGTGGAATTCCGCGTGTAGCAGTGAAATGCGTAG
ATATGCGGAGGAACACCGATGGCGAAGGCAGCCTCCTGGGATAACACTGACGCTCATGCACGAAAGCGTGGGGAGCAAACAGG
ATTAGATACCCTGGTAGTCCACGCCCTAAACGATGTCAACTAGCTGTTGGGGCCTTCGGGCCTTAGTAGCGCAGCTAACGCGTG
AAGTTGACCGCCTGGGGAGTACGGTCGCAAGATTAAAACTCAAAGGAATTGACGGGGACCCGCACAAGCGGTGGATGATGTGG
ATTAATTCGATGCAACGCGAAAAACCTTACCTACCCTTGACATGTCTGGAATTCCGAAGAGATTTGGAAGTGCTCGCAAGAGA
ACCGGAACACAGGTGCTGCATGGCTGTCGTCAGCTCGTGTCGTGAGATGTTGGGTTAAGTCCCGCAACGAGCGCAACCCTTGTC
ATTAGTTGCTACGAAAGGGCACTCTAATGAGACTGCCGGTGACAAACCGGAGGAAGGTGGGGATGACGTCAAGTCCTCATGGC
CCTTATGGGTAGGGCTTCACACGTCATACAATGGTCGGGACAGAGGGTCGCCAACCCGCGAGGGGGAGCCAATCCCAGAAACCC
GATCGTAGTCCGGATCGCAGTCTGCAACTCGACTGCGTGAAGTCGGAATCGCTAGTAATCGCGGATCAGCATGTCGCGGTGAA
TACGTTCCCGGGTCTTGTACACACCGCCCGTCACACCATGGGAGTGGGTTTTACCAGAAGTAGTTAGCCTAACCGTAAGGGGGG
CGATTACCACGGTAGGATTCATGACTGGGGTGAAGTCGTAACAA
B
Actinomycetales
Arthrobacter sp. IN13 strain IN13
Arthrobacter sp. 16.43
Arthrobacter sp. SS9.18
Arthrobacter humicola strain: S32113
Arthrobacter sp. FB24 complete genome
Arthrobacter oryzae S32118
Arthrobacter sp. SS9.32
Arthrobacter sp. SS6.4
Arthrobacter sp. S21020
Arthrobacter oryzae S32237
Arthrobacter sp. J3.62
Arthrobacter sp. S21106
Arthrobacter sp.Sulf-717
Arthrobacter oryzae strain KV-651 16S ribosomal RNA partial sequence
Arthrobacter sp. S21114 gene for 16S ribosomal RNA partial sequence
Arthrobacter sp. CN-1 16S ribosomal RNA gene partial sequence
Arthrobacter oryzae gene for 16S rRNA partial sequence strain: S32227
Arthrobacter sp. S21130 gene for 16S ribosomal RNA partial sequence
Arthrobacter oryzae gene for 16S rRNA partial sequence strain: S32226
Arthrobacter sp. SS9.19 16S ribosomal RNA gene partial sequence
Arthrobacter sp. B3116 16S ribosomal RNA gene partial sequence
Arthrobacter sp. FB24 strain FB24 16S ribosomal RNA complete sequence
Arthrobacter oryzae gene for 16S rRNA partial sequence strain: S32137
Arthrobacter sp. GW10-3 16S ribosomal RNA gene partial sequence
Arthrobacter oryzae gene for 16S rRNA partial sequence strain: S32112
gi|441089480|gb|KC160883.1|:1-1485 Arthrobacter sp. SS6.10 16S ribosomal RNA gene partial sequence
gi|116608677|gb|CP000454.1|:1678840-1680351 Arthrobacter sp. FB24 complete genome
gi|341925880|dbj|AB648958.1|:1-1485 Arthrobacter oryzae gene for 16S rRNA partial sequence strain: S32117
gi|76262450|gb|DQ157992.1|:1-1512 Arthrobacter sp. FB24 16S ribosomal RNA gene partial sequence
gi|339646894|gb|JF969174.1|:4-1492 Bacterium REGD2 16S ribosomal RNA gene partial sequence
gi|341925870|dbj|AB648948.1|:1-1485 Arthrobacter oryzae gene for 16S rRNA partial sequence strain: S32002
gi|116608677|gb|CP000454.1|:1145794-1147305 Arthrobacter sp. FB24 complete genome
gi|341925879|dbj|AB648957.1|:1-1486 Arthrobacter oryzae gene for 16S rRNA partial sequence strain: S32114
gi|76262443|gb|DQ157985.1|:1-1514 Arthrobacter sp. 16.6 16S ribosomal RNA gene partial sequence
gi|226816220|dbj|D84588.2|:1-1485 Arthrobacter sp. S22227 gene for 16S ribosomal RNA partial sequence
gi|341925887|dbj|AB648965.1|:1-1485 Arthrobacter oryzae gene for 16S rRNA partial sequence strain: S32220
gi|56462446|gb|AY833100.1|:1-1419 Actinobacterium EC5 isolate Ph37 16S ribosomal RNA gene partial sequence
gi|76262456|gb|DQ157998.1|:1-1512 Arthrobacter sp. J3.40 16S ribosomal RNA gene partial sequence
Arthrobacter spp.
gi|94995767|gb|DQ490461.1|:1-1490 Micrococcaceae bacterium KVD-unk-27 16S ribosomal RNA gene partial sequence
gi|378558097|gb|JQ396624.1|:1-1501 Arthrobacter sp. MN12-12 16S ribosomal RNA gene partial sequence
gi|402549862|gb|JX266387.1|:4-1491 Arthrobacter sp. B3027 16S ribosomal RNA gene partial sequence
gi|226816240|dbj|D84563.2|:1-1485 Arthrobacter sp. S21011 gene for 16S ribosomal RNA partial sequence
gi|402549856|gb|JX266381.1|:4-1491 Arthrobacter sp. B2102 16S ribosomal RNA gene partial sequence
gi|441422546|gb|KC236871.1|:1-1449 Arthrobacter sp. B3041 16S ribosomal RNA gene partial sequence
gi|341925882|dbj|AB648960.1|:1-1485 Arthrobacter oryzae gene for 16S rRNA partial sequence strain: S32126
gi|133751117|gb|EF459529.1|:3-1296 Actinobacterium KFC-24 16S ribosomal RNA gene partial sequence
Arthrobacter sp.
Arthrobacter sp.
Arthrobacter sp. S21003
Arthrobacter sp. B1033
Arthrobacter sp. S21004
Arthrobacter sp. J3.31
Arthrobacter sp. S22236
Arthrobacter humicola strain KV-653
Arthrobacter spp.
Arthrobacter sp. J3.46
isolate 16S rRNA gene
Arthrobacter sp. J3.16
Arthrobacter sp. J3.33
Actinobacterium EC5
Arthrobacter spp.
Arthrobacter spp.
Arthrobacter sp.
Actinobacterium MES16
Arthrobacter spp.
Arthrobacter sp. bD37(2011)
Bacterium W10
Arthrobacter spp.
Arthrobacter spp.
Arthrobacter globiformis
Arthrobacter spp.
Arthrobacter globiformis
Zhihengliuella alba
Arthrobacter sp. RS-33
Citricoccus sp. PL13f S6 Zhihengliuella alba
Psychrophilic marine bacterium PS32
Micrococcus lylae
Citricoccus sp. PL13f S6
Kocuria sp. IARI-R-30
Kocuria sp. IARI-R-30
Rothia sp. RV13
Arthrobacter isolate 16S rRNA sequence used for comparison:
AGAGTTTGATCCTGGCTCAGGATGAACGCTGGCGGCGTGCTTAACACATGCAAGTCGAACGATGATCCGGTGCTTGCACCGGG
GATTAGTGGCGAACGGGTGAGTAACACGTGAGTAACCTGCCCTTAACTCTGGGATAAGCCTGGGAAACTGGGTCTAATACCGG
ATATGACTCCTCATCGCATGGTGGGGGGTGGAAAGCTTTATTGTGGTTTTGGATGGACTCGCGGCCTATCAGCTTGTTGGTGA
GGTAATGGCTCACCAAGGCGACGACGGGTAGCCGGCCTGAGAGGGTGACCGGCCACACTGGGACTGAGACACGGCCCAGACTCC
TACGGGAGGCAGCAGTGGGGAATATTGCACAATGGGCGCAAGCCTGATGCAGCGACGCCGCGTGAGGGATGACGGCCTTCGGG
TTGTAAACCTCTTTCAGTAGGGAAGAAGCGAAAGTGACGGTACCTGCAGAAGAAGCGCCGGCTAACTACGTGCCAGCAGCCGC
GGTAATACGTAGGGCGCAAGCGTTATCCGGAATTATTGGGCGTAAAGAGCTCGTAGGCGGTTTGTCGCGTCTGCCGTGAAAGT
CCGGGGCTCAACTCCGGATCTGCGGTGGGTACGGGCAGACTAGAGTGATGTAGGGGAGACTGGAATTCCTGGTGTAGCGGTGA
AATGCGCAGATATCAGGAGGAACACCGATGGCGAAGGCAGGTCTCTGGGCATTAACTGACGCTGAGGAGCGAAAGCATGGGGA
GCGAACAGGATTAGATACCCTGGTAGTCCATGCCGTAAACGTTGGGCACTAGGTGTGGGGGACATTCCACGTTTTCCGCGCCGT
AGCTAACGCATTAAGTGCCCCGCCTGGGGAGTACGGCCGCAAGGCTAAAACTCAAAGGAATTGACGGGGGCCCGCACAAGCGG
CGGAGCATGCGGATTAATTCGATGCAACGCGAAGAACCTTACCAAGGCTTGACATGGACCGGACCGGGCTGGAAACAGTCCTT
CCCCTTTGGGGCCGGTTCACAGGTGGTGCATGGTTGTCGTCAGCTCGTGTCGTGAGATGTTGGGTTAAGTCCCGCAACGAGCGC
AACCCTCGTTCCATGTTGCCAGCGCGTAATGGCGGGGACTCATGGGAGACTGCCGGGGTCAACTCGGAGGAAGGTGGGGACGA
CGTCAAATCATCATGCCCCTTATGTCTTGGGCTTCACGCATGCTACAATGGCCGGTACAAAGGGTTGCGATACTGTGAGGTGG
AGCTAATCCCAAAAAGCCGGTCTCAGTTCGGATTGGGGTCTGCAACTCGACCCCATGAAGTCGGAGTCGCTAGTAATCGCAGA
TCAGCAACGCTGCGGTGAATACGTTCCCGGGCCTTGTACACACCGCCCGTCAAGTCACGAAAGTTGGTAACACCCGAAGCCGGT
GGCCTAACCCCTTGTGGGAGGGAGCTGTCGAAGGTGGGACTGGCGATTGGG-ACTAAGTCGTAACAAGGTA
Figure 1. Tree alignment of fragments of 16S rRNA genes. A) Achromobacter isolate 16S rRNA gene trimmed to 1460 bp (16S rRNA gene sequence based on gDNA sequencing) aligned against the top 100 hits in the GenBank NCBI database and other
Burkholderiales species (in order to root the tree). B) Arthrobacter isolate 16S rRNA gene trimmed to 1486 bp (16S rRNA gene sequence based on gDNA sequencing) aligned against the top 100 hits in the GenBank NCBI database and other Actinomycetales species (in order to root the tree).
Sequences were aligned using ClustalW and trees were constructed using MEGA5 software using Neighbor-Joining with Bootstrap of 1000 replicates (bootstrap values are shown on next to the tree nodes) (Felsenstein 1985, Saitou and Nei 1987, Tamura et al 2004, Tamura et al 2011). A Neighbor-Joining tree is commonly used for this kind of analysis (Micallef et al 2009). For the Arthrobacter alignment the node containing the isolates 16S rRNA gene sequence is resolved next to the main tree (black arrow).
The genome of Achromobacter xylosoxidans is 7.01 Mbp and that of Arthrobacter
sp. is 3.815 Mbp. These are minimum sizes because the contigs have not been closed and there may be missing DNA.
Felsenstein J (1985). Confidence limits on phylogenies: An approach using the bootstrap. Evolution; international journal of organic evolution 39: 783-791.
Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35: 3100-
3108.
Micallef SA, Shiaris MP, Colon-Carmona A (2009). Influence of Arabidopsis thaliana accessions on rhizobacterial communities and natural variation in root exudates. J Exp
Bot 60: 1729-1742.
Saitou N, Nei M (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular biology and evolution 4: 406-425.
Tamura K, Nei M, Kumar S (2004). Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A 101: 11030-11035.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular biology and evolution 28: 2731-2739.