Supporting information Materials and methods Strain isolation and
advertisement

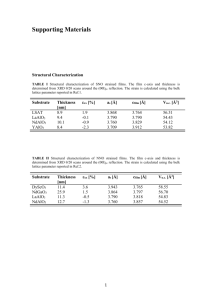
1 Supporting information 2 Materials and methods 3 Strain isolation and characterization 4 The DBHB-degrading strain was isolated from the surface soil collected from a manufacturing facility 5 producing halogenated aromatic compounds located in Jiangsu, China. The isolation procedure was 6 carried out in MM medium supplemented with 0.2 mM DBHB by the conventional enrichment culture 7 technique (Li et al. 2010). The bacterial strain EOB-17 showed the ability to degrade DBHB and was 8 isolated and purified. 9 Strain EOB-17 was identified based on its morphological, physiological and biochemical properties 10 with reference to Bergey’s Manual of Determinative Bacteriology and 16S rRNA gene sequence 11 analysis. Cell morphology was examined by light microscopy (Olympus; × 1000) using phase-contrast 12 optics and by transmission electron microscopy (H-7650, Hitach) using cells that had been grown for 13 16 hours at 30°C on LB medium. The growth of strain EOB-17 under anaerobic conditions was 14 determined using the GasPak Anaerobic Systems (BBL) according to the manufacturer’s instructions. 15 The genomic DNA of strain EOB-17 was extracted by high-salt precipitation (Miller et al. 1988). The 16 16S rRNA gene was amplified by a previously described PCR method (Thompson et al. 1997). The 17 nearly complete 16S rRNA gene sequence (1491 bp) was deposited in the GenBank database under the 18 accession number KR232560. Phylogeny was analyzed with MEGA version 4.0 software, and distance 19 was calculated using the Kimura 2 parameter distance model. An unrooted tree was built by the 20 neighbor-joining method (Kumar et al. 2008). The dataset was bootstrapped 1,000 times. 21 Supplementary Fig. S1 and Supplementary Fig. S2 22 References 23 Kumar S, Nei M, Dudley J, Tamura K (2008) MEGA: a biologist-centric software for evolutionary 24 analysis of DNA and protein sequences. Brief Bioinform 9:299-306 25 Li R, Zheng J, Wang R, Song Y, Chen Q, Yang X, Li SP, Jiang JD (2010) Biochemical degradation 26 pathway of dimethoate by Paracoccus sp. Lgjj-3 isolated from treatment wastewater. Int Biodeter 27 Biodegr 64:51-57 28 29 30 Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acid Res 16:1215 Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows 31 interface: flexible strategies for multiple sequence alignment aided by quality analysis 32 tools. Nucleic Acid Res 25:4876-4882 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 Supplementary Table 1 Sequence comparison of deduced DBHB-degrading genes in Delftia sp. 62 EOB-17 Gene (accession Proposed product No.) bhbA3 No. of amino Homologous protein (GenBank Identity acids accession No.) and host % Reductive BhbA (YP_007878394), Comamonas 1071 (KR232561) dehalogenase bhbB3 Extracytoplasmic 99 sp. 7D-2 BhbB (YP_007878395), Comamonas 337 (KR232562) binding receptor bhbF3 4-hydroxybenzoate (KR232563) 3-monooxygenase 99 sp. 7D-2 4-hydroxybenzoate 391 3-monooxygenase (WP_016446365), 99 Delftia acidovorans protocatechuate protocatechuate 3,4-dioxygenase bhbD3 3,4-dioxygenase 121 (WP_027016029), 93 (KR232564) alpha subunit Comamonas composti protocatechuate protocatechuate 3,4-dioxygenase bhbE3 3,4-dioxygenase 289 (WP_043822444), (KR232564) beta subunit 63 64 65 66 67 68 69 70 71 72 73 74 75 Delftia sp. RIT313 99 76 Supplementary Fig. S1 Transmission electron micrograph of a negatively stained cell of strain 77 EOB-17 shows a rod (0.7-0.9 μm × 1.7-2.2 μm) shape with polar flagella. Bar, 1.0 µm. 78 Supplementary Fig. S2 Neighbor-joining phylogenetic tree based on 16S rRNA gene sequences 79 shows the relationship of strain EOB-17 and its related taxa. Bootstrap values (expressed as 80 percentages of 1,000 replications) > 50% are shown at branching points. Bar, 0.005 substitutions per 81 nucleotide position. 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 Supplementary Fig. S1 127 128 129 Supplementary Fig. S2