tpj12050-sup-0011-Supporting Information legends
advertisement

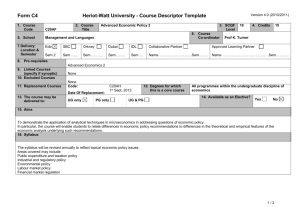
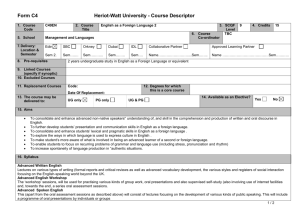
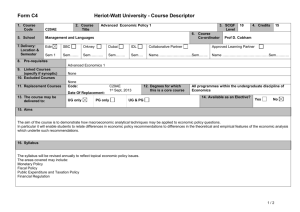
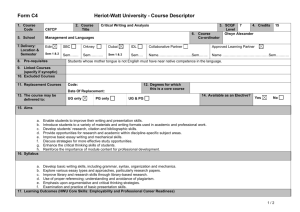
Supporting Information legends Figure S1. PSKR1 but not PSKR2 and PSY1R expression is enhanced after bacterial infection and PAMP treatment. Mean relative expression values from microarray data from AtGenExpress with standard errors from three replicate arrays after the indicated Pseudomonas infections (a) or PAMP treatments (b) are shown Figure S2. Relative expression of PSK1, PSK2, PSK3, PSK4 and PSK5 from microarray data from AtGenExpress. Mean expression values and standard error from three replicate arrays after Pseudomonas treatment (a) or PAMP treatment (b). Figure S3. Relative expression of PSY1, and its two paralogs At3g47395 and At2g29995 from microarray data from AtGenExpress. Mean expression values and standard error from three replicate arrays after Pseudomonas treatment (a) or PAMP treatment (b). Figure S4. Mutants lacking PSKR1 are more resistant to the bacterial pathogen Pseudomonas syringae pv. tomato DC3000. Bacterial growth was monitored at 0 to 4 days post inoculation with 104 cfu/ml Pto DC3000 in fiveweek-old plants of the indicated genotypes. Mean values of the number of bacteria are presented +/SEM of at least 6 biological replicates, asterisks represent significant differences from Col-0 (*p<0.5, **p<0.01, Student’s t-test) (a). Plants were photographed 5 days after spray inoculation with 108 cfu/ml Pto DC3000 (b). Leaves of spray inoculated plants were subjected to trypan blue staining 5 days after infection (c). Scale bar represents 0.2 mm. Representative results of three experiments are presented. Figure S5. pskr2 does not affect pskr1 or psy1r Alternaria brassicicola susceptibility phenotypes in double and triple mutants. Disease symptoms were monitored at 7, 10 and 13 days after inoculation with 105 spores/ml. Mean values are presented +/- SEM of at least 15 replicates. Asterisks represent significant differences from Col-0 (*p<0.5, **p<0.01, Student’s t-test) (a). Pictures of representative leaves were taken 13 days after inoculation (b). Leaves were subjected to trypan blue staining for visualization of mycelial growth 10 days after inoculation (c). Scale bar represents 0.2 mm. Figure S6. pskr2 does not affect pskr1 or psy1r Pseudomonas syringae pv. tomato resistance phenotypes in double and triple mutants. Bacterial growth was monitored at 0 to 4 days post inoculation with 104 cfu/ml Pto DC3000. Mean values are presented +/- SEM. Asterisks represent significant differences from Col-0 (*p<0.5, ***p<0.001, Student’s t-test). Figure S7. Expression of GFP-tagged PSKR1 partially complements the Pto DC3000 resistance and Alternaria brassicicola susceptibility phenotypes of pskr1-3/pskr2/psy1r plants. (a) Plants were inoculated with 104 cfu/ml Pto DC3000, growth was monitored at 0 to 4 days after infection. Mean values of bacterial numbers are presented +/- SEM of at least 6 biological replicates. (b) Plants were treated with 105 spores/ml Alternaria brassicicola. Disease indices were rated at days 7 to 13. Mean values are presented +/- SEM of at least 15 replicates. Bars with different letters are significantly different based on one-way ANOVA (p<0.05, Student’s t-test). (c) Alternaria brassicicola treated plants were photographed 13 days after inoculation. Figure S8. PAMP responses are altered in tpst mutants. Adult plants were infiltrated with or without 100 nM flg22 for 24 hours and (a) callose deposition was visualized by aniline blue staining. Signal intensity was plotted as mean values +/- SEM from 10 replicates. Asterisks represent significant differences from Col-0 (***p<0.001, Student’s t-test) (b). Biomass of flg22 induced inhibition of seedling growth was presented as mean values +/- SEM from 6 replicate samples containing 8 seedlings each. Asterisks represent significant differences from Col-0 (***p<0.001, Student’s t-test) (c). For FRK1 gene expression analysis, qRT-PCR experiments were performed on cDNA generated from three independent biological replicates. Expression values were normalized to EF1α and then expressed as a ratio to Col-0 and presented as fold induction. Bars represent mean ratios +/- SEM (d). Scale bar represents 0.2 mm. Representative results of three experiments are presented. Figure S9. PSKα treatment leads to enhanced root growth in Col-0 seedlings. To test that our preparation of PSKα was active seedlings were grown on 1/2 MS media containing either PSKα or buffer only. Root length measurements were taken 6 days after transfer. Bars represent mean values of 30 replicate seedlings +/- SEM. Asterisks represent significant differences from Col-0 (***p<0.001, Student’s t-test). Table S1. List of primers used in PCR experiments.