tpj12051-sup-0011-Supporting information legends
advertisement

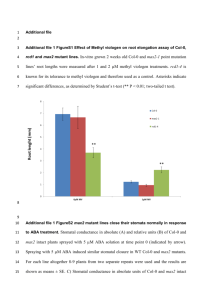
1 Supporting Information legends 2 3 Figure S1. MeJA Response of AtMYB44 overexpressing and atmyb44 knockout 4 plants. 5 (a) Root lengths of Arabidopsis Col-0, AtMYB44 overexpression plants (OX18) and 6 atmyb44 knockout plants were measured. Data represent the mean root length from 7 fifty seedlings. Error bars represent standard deviation. 8 (b) Root hair density was quantified from Arabidopsis Col-0, AtMYB44 9 overexpression plants (OX18, OX21) and atmyb44 knockout plants. The values 10 represent the mean of fifteen seedlings. Error bars represent standard deviation. 11 Statistical significance of the measurements was determined using a t-test (*P<0.01) 12 by comparison with the value of Col-0. 13 14 Figure S2. Resistance of npr1-1 and NahG plants against the biotrophic pathogen 15 Pst DC3000. 16 Four-week-old Arabidopsis Col-0, npr1-1 and NahG plants were inoculated with Pst 17 DC3000. Bacterial growth in leaves was determined one (open bars) or three days 18 (closed bars) after inoculation. Statistical significance of the measurements was 19 determined using a t-test (*P<0.05) by comparison with the value of Col-0. Data 20 represent the mean values of eight independent plants and error bars represent 21 standard deviation. 22 23 Figure S3. Expression of SA signaling genes in AtMYB44 overexpressing and 24 atmyb44 knockout plants. 25 Expression of SA signaling genes was determined by RT-PCR with a limited number 26 of cycles (25 cycles, Method S1) and PCR product was visualized by ethidium 27 bromide staining. 28 29 Figure S4. Transcriptional activation domain assay of AtMYB44. 30 (a) Various fragments of AtMYB44 were recombined with the GAL4 binding domain 31 as shown in the diagram. 32 (b) Transcriptional activity was measured by –galactosidase assay (Method S1). 1 33 Data represent mean values of three independent measurements. Error bars 34 represent standard deviation. 35 36 Figure S5. Direct binding of AtMYB44 to the WRKY70 promoter in vivo. 37 (a) Interaction of AtMYB44 with WRKY70 promoter was tested by yeast one-hybrid 38 (Y1H) assay. WRKY70 promoter sequence of -328 to -299 containing the core 39 binding sequence (RE44) or its mutant version (mRE44) was repeated 4 times and 40 combined with HIS3 for the Y1H assay. A cDNA encoding the whole AtMYB44 41 protein was fused with the activation domain as bait. 42 (b) Structure of WRKY70 indicating regions amplified by PCR (top). Open boxes at 43 sites 1, 2 and 4 represent the core AtMYB44 binding sequence. PCR primer sets 44 binding to another promoter region (Site 3), CDS region (Site 5) or 3’-UTR region of 45 WRKY70 (Site 6) were also employed for negative controls. Fragmented chromatin 46 DNA of Arabidopsis Col-0 and AtMYB44-GFP overexpressing plants were 47 immunoprecipitated with anti-GFP and analyzed by PCR (bottom left). Chromatin 48 DNA before immunoprecipitation was PCR amplified as an input control. 49 50 Figure S6. SA-mediated suppression of JA response in atmyb44 knockout mutant 51 plants. 52 Two-week-old Arabidopsis plants were treated with combination of SA and JA. 53 Treated seedlings were harvested 6 hour after treatment. Total RNA was analyzed by 54 Northern blot and rRNA was visualized by ethidium bromide staining as a loading 55 control. 56 57 Figure S7. Expression of AtMYB44 to Pst DC3000. 58 Four-week-old Arabidopsis Col-0 plants were treated with Pst DC3000. rRNA is 59 shown for checking equal loading. 60 61 Figure S8. Induction of WRKY70 and PR1 by MeJA. 62 Arabidopsis Col-0 and coi1-1 mutants were treated with 50 μM of MeJA for the 63 indicated number of hours and JA-responsive genes were analyzed by Northern blot. 64 rRNA was visualized by ethidium bromide staining as a loading control. 2 65 66 Table S1. Primers used in Northern blot analysis, ChIP PCR and subcloning. 67 68 Methods S1. Supplemental experimental procedures. 3