Assignment 1
advertisement

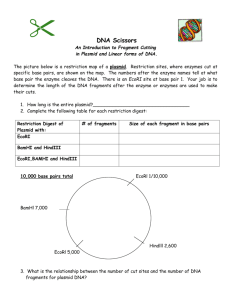
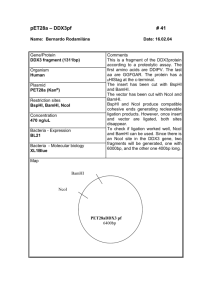
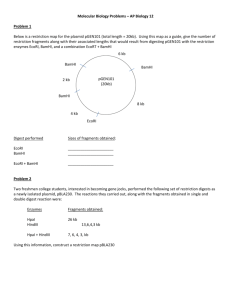
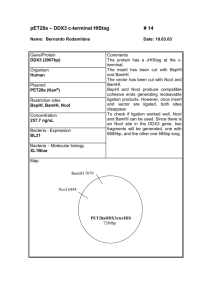
Assignment 1 Dilutions and Concentrations Only submit the answers to one digit after the decimal, you do not need to show your calculations 1. You dilute 175 mL of 1.6 M LiCl to 1.0 L, what is the new concentration of the solution? 2. You need to make 10.0 L of 1.2 M KNO3. What molarity would the potassium nitrate stock solution need to be if you were to use only 2.5 L of it? 3. How many milliliters of 5.0 M copper sulfate solution must be added to 160 mL of water to achieve a 0.30 M copper sulfate solution? 4. A 40.0 mL volume of 1.80 M Fe(NO3)3 is mixed with 21.5 mL of 0.808M Fe(NO3)3 solution. Calculate the molar concentration of the final solution. 5. To 2.00 L of 0.445 M HCl, you add 3.88 L of a second HCl solution of unknown concentration. The new solution is 0.974 M. What was the molarity of the second HCl solution? 6. 1L of a solution is prepared by dissolving 125.6 g of NaF (MW: 42) in it. If you took 180 mL of that solution and diluted it to 500 mL, what would be the molarity of the resulting solution? 7. What is the molar concentration of chloride ions in a solution prepared by mixing 100.0 mL of 2.0 M KCl with 50.0 mL of a 1.50 M CaCl2 solution? 8. The A260nm of a DNA solution is 0.12. What volume is required to have 30ng of DNA? 9. You have a DNA solution at a concentration of 6ng/µL and wish to prepare 50µL of a solution containing 30ng of DNA. Given that your P20 pipette is broken, explain how you would prepare this solution. 10. You have a DNA solution at a concentration of 0.4µg/µL. How would you prepare a 1mL solution of this DNA which would give an absorbance at 260nm of 0.04? 11. What would be the percent by mass (% m/m) of a solution made by dissolving 54 g of AgNO3 in 128 g of water? 12. If you make a solution by adding water to 75 mL of ethanol until the total volume of the solution is 375 mL, what’s the percent by volume (% v/v) of ethanol in the solution? 13. What volume of a 2.73 M NaOH solution would you need to prepare 142 mL of a 0.540 M solution? 14. How many of grams of KCl are in 3L of a 2% (v/v) solution. (Density of KCl :1.2) Restriction Mapping 1. The linear 12 Kbp DNA fragment shown below has cleavage sites for BamHI and EcoRI. The numbers indicate the distance in kilobases. Complete the table to indicate the fragment sizes which would be generated following each of the indicated digests. B E B 2 4 6 Enzyme digest E 8 10 Fragment sizes BamHI EcoRI BamHI + EcoRI 2. What fragment sizes could be generated from a BamHI partial digest? Do not include fragment sizes resulting from a complete digest. 3. Draw all possible maps, excluding the one shown above, which could correspond to the results indicated in the above table for the BamHI digest alone. 4. The circular DNA molecule shown below has cleavage sites for BamHI and EcoRI. Complete the table to indicate the fragment sizes which would be generated following each of the indicated digests. Enzyme digest Fragment sizes BamHI EcoRI BamHI + EcoRI 5. What fragment sizes could be generated from a BamHI partial digest? Do not include fragment sizes resulting from a complete digest. 6. Is this the only possible map according to the results presented in the above table? If not draw another possible map. 7. A 8.9 kb circular plasmid is digested with three restriction enzymes, EcoRI, BamHI and HindIII, individually and in combination, and the resulting fragment sizes are determined by means of electrophoresis. The results are as follows: EcoRI BamHI HindIII EcoRI + BamHI EcoRI + HindIII BamHI + HindIII BamHI + EcoRI + HindIII Draw a possible restriction map based on these results. 8.9 kb 6 kb. 2.9 kb 8.9 kb 6 kb, 2.4 kb, 0.5 kb 7.4 kb, 1.5 kb 5 kb, 2.9 kb, 1 kb 5 kb, 2.4 kb, 1 kb, 0.5 kb