Ballarino Ponzano 2014
advertisement

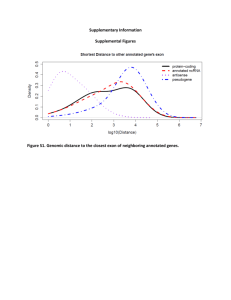
Transcript expression profiling identifies novel long non-coding RNAs (lncRNAs) modulated during murine myogenic differentiation. Monica Ballarino1, Valentina Cazzella1, Mariangela Morlando1, Andrea Cipriano1, Lavinia Bisceglie1, Anna Tramontano2,3 and Irene Bozzoni1,3 Department of Biology and Biotechnology ‘‘Charles Darwin’’ Department of Physics 3 Center for Life Nano Science @Sapienza, Istituto Italiano di Tecnologia Sapienza University of Rome, P.le A. Moro 5, 00185 Rome, Italy 1 2 Forthcoming ambition in the RNA field is to integrate long non-coding RNAs (lncRNAs) into the molecular circuitries controlling cell differentiation. Along this direction, we recently discovered linc-MD1, a cytoplasmic lncRNA which cross-regulates myogenic mRNAs by competing for microRNA (miRNA) binding. These findings opened the intriguing possibility that lncRNA-based mechanisms might influence different sets of transcripts during the achievement of myogenic programs. Even though large-scale efforts have been done to list lncRNAs expressed in muscle cells (myolncRNAs), the current knowledge is still partial and limited by the multiplicity of annotation criteria. Here, we expanded the existing collection of murine myo-lncRNAs by using a strand-specific RNA sequencing approach on growing C2C12 myoblasts and differentiated myotubes. We found a subset of nuclear and cytoplasmic lncRNAs which expression is dynamically ordered during differentiation. Semi-quantitative RT-PCR analyses also revealed the existence of splicing isoforms switching during differentiation and restricted to different subcellular compartments. Moreover, the analysis of mouse tissues identified several tissue-specific lncRNAs and lncRNAs whose expression was altered in mdx dystrophic muscles, suggesting a potential impact of the newly identified transcripts in muscle pathophysiology. Among the differentially expressed lncRNAs interestingly we found a new cytoplasmic lncRNA transcribed from miR-31 genomic locus and specifically expressed in C2C12 growing myoblasts. Because of the involvement of miR-31 in both muscle physiology and pathology, the study of lnc-31 function has been prioritised and preliminary data on the effect on lnc31 overespression and depletion will be presented. Overall, our data lay the foundation for further investigations enlarging the landscape of RNA molecules involved in the acquisition of muscle identity.