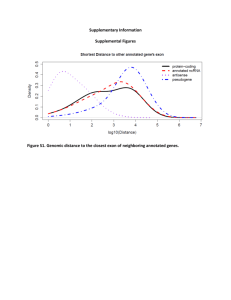
tpj12679-sup-0026-MethodsS1
advertisement

Supplemental methods
Ribo-minus and non-polyA RNA sequencing
We first sequenced total RNAs from the mixed tissues (inflorescences, rosette
leaves, cauline leaves and stems) of Arabidopsis. Tissues and seedlings were
frozen in liquid nitrogen and immediately used to isolate total RNAs (QIAGEN
RNeasy Plant Mini Kit), with a further step of DNase treatment. The RNAs
were then quantified using NanoDrop 1000. The same amount of tissue RNAs
were mixed. Ribosomal RNAs were then deleted using the RiboMinus™ Plant
Kit, repeated three times. RNAs were then fragmented and enriched by
agarose gel at ~330 nt. A strand-specific cDNA library was constructed by
ligating different adapters according to the SMART library construction method
(Levin et al., 2010), and then sequenced by Illumina HiSeq 2000 using the 100
nt paired-end sequencing protocol.
We then sequenced nine poly(A)- RNAs from seedlings treated with
different stress conditions at different times (Table S1). The poly(A)- RNAs
were collected by removing poly(A)+ RNAs four times using the Oligotex
mRNA Mini Kit (QIAGEN). Then rRNAs were removed from poly(A)- RNAs
three times as above. Fragmentation and enrichment of RNAs after poly(A)+
RNAs and rRNAs removal were the same as described for the mixed tissue
RNA sequencing. Fragments (200~500 nt) were excised from the gel and
purified for sequencing. We used the SMART™ cDNA Library Construction Kit
for strand-specific cDNA library construction. The 100 nt single-end
sequencing protocol of Illumina HiSeq 2000 was selected.
Transcriptome reassembly and filtering for predicted lncRNA transcripts
For the mixed-tissue total RNA-seq, the first four and the last five nucleotides
of raw reads were trimmed; for poly(A)- RNA-seq data, the first three
nucleotides of raw reads were trimmed because of poor quality using the
FASTX-toolkit. Reads were then mapped to TAIR10 rRNA sequences by
Bowtie (v 0.12.9) with one mismatch, and retained reads were further mapped
to the TAIR10 genome using TopHat (v 2.0.8) with two mismatches.
Transcriptomes were re-assembled using the Cufflinks and Cuffmerge
programs following the pipeline (Trapnell et al., 2012). Then, we performed
several stringent filters to clean potential noise: [1] we filtered the transcripts if
they overlapped TAIR10 annotated coding genes or noncoding RNAs with
more than one nucleotide on the same strand, [2] we filtered the transcripts if
they were able to align with specific types of canonical ncRNAs in miRBase,
Plant snoRNAdb, tRNAdb or Rfam (Brown et al., 2003, Burge et al., 2013,
Juhling et al., 2009, Kozomara and Griffiths-Jones 2011, Trapnell et al., 2012)
using the Blast program (default cutoff), [3] we used INFERNAL to scan the
Arabidopsis genome for homologs of structured ncRNAs in Rfam (E <= 0.01
as cutoff), and filtered overlapping transcripts, [4] we calculated the CPC
scores using the CPC program (default parameters), and filtered ncRNA
transcripts with higher coding potential (CPC score > 0) (Kong et al., 2007),
and [5] we filtered several transcripts that aligned with mitochondria or
chloroplast sequences using the BLASTn program (default cutoff). We have
kept 409 assembled transcripts that are overlapping with the annotated
lncRNAs, and performed the filtering steps in Figure 1. We found 18 transcripts
can be aligned to the known sequences of canonical ncRNAs annotated in
miRBase, Plant snoRNAdb, or tRNAdb, 6 transcripts have homologs in Rfam,
and 35 transcripts have coding potential score (CPC score) larger than 0.
Overall, 85.6% of the lncRNA passed the filters. The false negative rate is
14.4%, if we assume that all the annotated lncRNAs are truly functional.
However, we need to note that most of the annotated lncRNAs in the database
have not been well validated by solid experiments.
Gold-standard set of annotations
The gold-standard annotations of Arabidopsis thaliana genome were
downloaded from TAIR database version 10 (Lamesch et al., 2012). We first
defined the genic regions using the annotation of coding genes, all kinds of
known ncRNAs, pseudogenes, and transposable elements. The regions that
are 500 nt from the genic regions were defined as intergenic regions. The
known ncRNAs include 899 canonical ncRNAs (4 rRNAs, 631 tRNAs, 71
snoRNAs, 299 miRNAs, 13 snRNAs and 9 siRNAs) and 469 lncRNAs
(annotated as “other RNA” in TAIR10, filter out siRNA primary transcripts and
those shorter than 200 nt).
For model training and testing, the entire Arabidopsis thaliana genome
was split into 4,765,850 small bins (each 100 nt), and each two neighboring
bins have 50 nt overlap. If a genomic bin has more than 50 nt overlapped with
known ncRNAs in gold-standard set, it was defined as a ncRNA bin; if a bin
has more than 90 nt overlapped with CDS, UTR or intergenic region, it was
annotated correspondingly (Lu et al., 2011).
High-throughput data process
We have curated 147 high throughput sequencing and microarray data in our
integrative model (Table S2A). Twenty-six sets of poly(A)+ RNA sequencing
data were collected, sixteen were treated under various stress conditions
(SRA: SRP000935) (Filichkin et al., 2010), the remaining were produced from
different generations of Col-0 wild-type plants at normal condition (ENA:
ERP000902) (Becker et al., 2011). Thirteen sets of small RNA sequencing
data were from three groups, with GEO accessions GSE10967 (Lister et al.,
2008), GSE11070 (Gregory et al., 2008), and GSE11094 (German et al.,
2008). For these RNA-seq data, raw reads (fastq format) were first mapped by
Bowtie with the parameters -m 10 -v 1(-v 0 in small RNA-seq). The normalized
value (RPKM, reads per kilobase per million) was then assigned to each
genomic bin (every 100nt of the genome) by DEGseq package (Wang et al.,
2010). The maximum bin value among multiple samples was taken as the bin
score if sequenced by the same technology (e.g. poly(A) enriched).
We also collected seventy-eight poly(A)+ RNA tiling array data (GEO:
GSE13584) from (Zeller et al., 2009) and six total RNA tiling array data from
(Laubinger et al., 2008). Normalized signals for probes were calculated by R
package AffyTiling, and the average intensity of probes was assigned to each
bin (every 100nt of the genome). The feature score of each bin was counted as
done with sequencing data. We defined a set of unexpressed intergenic
regions as a negative control, whose expression are lower than the mean
expression of all genomic elements in all of the RNA-seq and array samples
(Lu et al., 2011).
The ChIP-seq data for histone modification was GSE28398 (Luo et al.,
2012). Histone modifications were conducted from raw reads, after bowtie
mapping (same parameters with small RNA-seq), we used MACS14 to convert
mapped reads to background normalized signals, and assign averaged signals
on each bin. Peaks for histone modification were identified with default p-value
of cutoff 1e-05, if the summit of peak located in 1.5 kb upstream of a TSS
(transcription start site) or gene body region, the genomic element (mRNA or
lncRNA) was defined as the target of this modification (Charron et al., 2009,
Zhou et al., 2010).
ChIP-seq data for transcription factor binding sites were from several
public datasets (used in lncRNA characterization), with GEO IDs GSE35059,
and GSE35315 (Oh et al., 2012). The TF signals were calculated from five
ChIP-seq data sets, similar to histone modification. Peaks were also called by
using MACS14 with default p-value cutoff 1e-05, if the summit of peak located
in 1 kb upstream or 100 bp downstream of a TSS (transcription start site), the
genomic element was defined as the target of this TF (Charron et al., 2009,
Zhou et al., 2010). BS-seq data for DNA methylation data was also from SRA:
ERP000902 (Becker et al., 2011). DNA methylation analysis was performed
using Bismark software with default parameters (Liao et al., 2011). The
probability of methylated cytosine of CpG, CHH, CHG context was calculated,
and sum of these probabilities on each bin was considered as the methylation
feature score. The DNA methylation target was defined if the genomic element
(mRNA, known ncRNA and predicted lncRNA candidates) contained
methylation sites (probability of methylated cytosine above 80%) (Chuang et
al., 2012).
Calculations of sequence and structural features
We calculated seven sequence and structure features (GC content, DNA
sequence conservation, protein sequence conservation, coding potential, RNA
secondary structure stability, RNA structure conservation, homologs in Rfam)
for each genomic bin (every 100nt of genome) (Table S2B). DNA conservation
was measured with BLASTn using default parameters; the genome sequences
of 31 plant species downloaded from PlantGDB were used as library database.
The maximum bit score was used as the feature score. Protein conservation
was identified using BLASTx in a similar manner. The coding potential of each
bin was measured by RNAcode with default parameters (Washietl et al., 2011).
RNA secondary structure stability of bins was calculated by RandFold program
(Bonnet et al., 2004), with 1000 times of dinucleotide random shuffling, and the
p-value was used as the feature score. The RNA structure conservation value
was denoted by SCI (structure conservation index) score, which was
calculated by RNAz based on the multiple sequence alignments of A.thaliana,
A. lyrata, T. halophile, C. papaya and C. clementine (downloaded from VISTA
database). We assigned the minimum RNA structure stability score (RandFold
p-values) and the maximum SCI score to the lncRNAs from overlapped bins.
We used INFERNAL program (Nawrocki et al., 2009) to scan Arabidopsis
genome, and identified all the homologs in Rfam. Then we assigned 1 or 0 to
each bin according to whether it overlaps with the homologs (overlap at least
50 nt).
Integrative model based on multiple features
We predicted a ncRNA score for the whole Arabidopsis genome with a
supervised machine learning method, incRNA (Gerstein et al., 2010, Lu et al.,
2011). We curated 147 public datasets, which included 26 sets of RNA-seq
data for poly(A)+ RNA, 78 sets of microarray data for poly(A)+ RNA, ten sets of
ChIP-seq data for histone modification, and 14 sets of bisulfite-seq data for
DNA-methylation (Table S2). In addition, we calculated seven computational
scores (i.e., GC content, DNA conservation, protein conservation, RNA
structure stability, RNA structure conservation, coding potential, and homologs
in Rfam) over the entire Arabidopsis genome (see below). Using these
high-throughput data and computational scores as input features, we built a
model for ncRNAs (canonical and annotated lncRNAs), using CDSs (coding
sequences), and unexpressed intergenic regions as negative controls. The
gold-standard training and testing sets were based on the annotations of TAIR
10. Based on the gold-standard sets, we used 2/3 of the them for training, and
the remaining 1/3 for independent validation, the model performance is shown
in Figure S3a. The incRNA model (classifier: Random Forest) predicted the
probabilities of every bin of the Arabidopsis genome (100 nt, two adjacent bins
have 50 nt overlapped) to be ncRNA (either canonical ncRNA or lncRNA). We
call this probability the ncRNA score.
Feature score comparisons
We calculated 25 feature scores for each genomic bin (Table S2). By
assigning these scores to coding or noncoding transcripts, we compared their
signal intensities for different groups of transcripts. For the features that tend to
carry local effects (i.e., DNA conservation, structure features, and histone
modification features), we calculated the maximum bin scores for each
transcript. For the other features (e.g. RNA-seq signals), we calculated the
average bin score for each transcript, and used these scores to measure
signal intensities of transcripts. For comparison between lncRNAs and
negative controls, we used the Mann-Whitney test to determine if the
differences were significant (p-value< 0.05).
Genomic locations of lncRNAs
The genomic locations of the lncRNAs were summarized and compared in
Figure 2c and Figure S6. If a lncRNA overlapped with small RNA-seq reads
(the same smRNA-seq data used in the model) with more than one nucleotide,
we classified it as small RNA (smRNA) precursor. For the other lncRNAs, if the
overlapped element is pseudogene or transposable element (TE), we
classified the lncRNA correspondingly. Antisense lncRNA means more than 50%
length of a lncRNA overlapped with known coding transcripts on the opposite
strand. Intergenic lncRNAs are located 500 nt away from coding transcripts or
annotated ncRNAs (Liu et al., 2012), and cis lncRNAs are located within 500 nt
from them without any overlaps.
Sequence conservation in multiple plant species
A
phylogenetic
tree
was
constructed
based
on
divergence
time
(http://www.timetree.org/), and a neighbor-joining tree was built using
MEGA5.0 (Tamura et al., 2011) (Figure S10). DNA conservation between
Arabidopsis thaliana and 16 other organisms was measured using BLASTn.
Paired t-tests were performed to determine if there were significant differences
between lncRNAs and coding genes, or intergenic regions.
Stress specificity score
We adapted the tissue specificity score method (Cabili et al., 2011) to calculate
a stress specificity score measuring the expression specificity responding to
stress. This method uses Jensen-Shannon divergence to measure the
distance between two expression patterns: 1, a transcript’s expressions across
n stress conditions, 𝑒; and 2, a predefined extreme case of expression pattern,
𝑒 𝑠 (only expressed under stress condition 𝑠), 𝑒 𝑠 = {
1, 𝑖𝑛 𝑠𝑡𝑟𝑒𝑠𝑠 𝑐𝑜𝑛𝑑𝑖𝑡𝑖𝑜𝑛𝑠
.
0, 𝑜𝑡ℎ𝑒𝑟 𝑐𝑜𝑛𝑑𝑖𝑡𝑖𝑜𝑛𝑠
The distance is
𝐽𝑆𝑑𝑖𝑠𝑡 (𝑒, 𝑒 𝑠 ) = √𝐽𝑆(𝑒, 𝑒 𝑠 ) ,
(1)
where the JS divergence of two probability distributions, 𝑝1 ,𝑝2 , is defined to be
𝑝1 +𝑝2
𝐽𝑆(𝑝1 , 𝑝2 ) = 𝐻 (
2
)−
𝐻(𝑝1 )+𝐻(𝑝2 )
2
,
(2)
where H is the entropy of a discrete probability distribution as follows
𝑝 = (𝑝1 , 𝑝2 … 𝑝1𝑛 ), 0 ≤ 𝑝𝑖 ≤ 1 𝑎𝑛𝑑 ∑𝑛𝑖=1 𝑝𝑖 = 1 ,
(3)
Then, the stress specificity of the transcript’s expression pattern, 𝑒, across n
stress conditions with respect to stress 𝑠 can be defined as
𝐽𝑆𝑠𝑝 (𝑒|𝑠) = 1 − 𝐽𝑆𝑑𝑖𝑠𝑡 (𝑒, 𝑒 𝑠 ) ,
(4)
Lastly, the stress specificity score of the transcript is defined as the maximal
stress specificity score across all n stress conditions such that
𝐽𝑆𝑠𝑝 (𝑒) = 𝑎𝑟𝑔𝑚𝑎𝑥𝑠 𝐽𝑆𝑠𝑝 (𝑒|𝑠), 𝑠 = 1 … 𝑛.
Differential expression analysis
(5)
To find differentially expressed lncRNAs in plants exposed to different stress
conditions, we assigned read counts of the poly(A)+ and poly(A)- lncRNAs with
poly(A)+ RNA-seq and poly(A)- RNA-seq data separately using the DEGseq
package (Wang et al., 2010).We normalized the raw read counts against the
total mapped reads in each sample, and then used the MA-plot based method
with a random sampling model to calculate the significance (p-values) and fold
change of differential expression Different time points of the same stress
treatment were merged together in the analysis. We used the fold change to
measure the expression variations between stresses and control; k-means
clustering was performed and the resulting heat maps are shown in Figure S11.
The p-value of 0.05 and a two-fold change was set as the differential
expression cutoff. When comparing the ratios of differentially expressed
transcripts between lncRNAs, and coding genes, we used the χ2 test and with
a stringent cutoff (p-value < 0.01).
Co-expression network and gene ontology enrichment analysis
We built a stress-related co-expression network based on 12 poly(A)+
RNA-seq data (Filichkin et al., 2010), and nine poly(A)- RNA-seq data that we
sequenced from plants exposed to five different stress conditions. The
poly(A)+ and poly(A)- lncRNAs’ RPKMs were calculated using the
corresponding data. In the input gene list, we included all the GO annotated
coding genes with lncRNAs. We retained only genes and lncRNAs whose
maximum RPKM among multiple samples was above the 75% percentile of all
expression values. The Pearson correlation coefficient (Pcc) was calculated to
define co-expressed pairs (Pcc>0.95 as cutoff). The Bonferroni multiple test
correction was introduced to each co-expressed pair, and a q-value of 0.01
was set as the cutoff. To annotate the co-expressed lncRNAs, we removed
lncRNAs with less than three neighbors. The GO enrichment analysis was
performed on the known classified neighbors using AgriGO (Du et al., 2010).
Sequence and structural motif search
Conserved sequence motif searches in a group of lncRNAs were carried out
by MEME (Bailey et al., 2009), with motif width constrained to 4 to 12
nucleotides, which is the common RBP binding size. The significance
threshold was set to an E-value of 0.05.
For the lncRNAs selected by the co-expression network, we predicted the
conserved structural motifs in grouped lncRNAs using the RNApromo (Rabani
et al., 2008) program ‘rnamotifs08_motif_finder.pl’ (p-value < 0.001). Then, we
used the predicted motifs to scan for similar structural motifs in all lncRNAs
studied here. Before scanning, we refined a stringent set of lncRNAs: we
calculated the structure conservation index (SCI) by RNAz (Gruber et al., 2010)
for five closely related plant genomes (Arabidopsis thaliana, Arabidopsis lyrata,
Carica papaya, Thellungiella halophile, Citrus clementina) for all lncRNAs, and
only retained the conserved lncRNAs (SCI >0.5). The RNApromo program
‘rnamotifs08_motif_match.pl’ was used for scanning, with a likelihood score >0
and a FPR=0.05.
To further verify the motifs, the background sequences were taken from
more than 1000 bp upstream of the lncRNAs, with the same lengths (Zhong et
al., 2010). For sequence motifs detected by MEME, we used MAST to search
them in the background sequences. The default cutoff (p-value<0.0001) was
used for motif match. We calculated the enrichments by comparing the number
of matched motifs in lncRNAs to that in the background sequences, and used
Fisher’s exact test to evaluate the statistical significance. For structural motif,
we used a RNApromo program (Rabani et al., 2008) “rnamotifs08_motif
_match.pl” to search the structural motifs (detected by “rnamotifs08
_motif_finder.pl”) in both lncRNAs and the background sequences. We used
the matched structural motifs (log-likelihood score >0) to calculate the
enrichment (fold-change). Then we used Mann-Whitney test to measure the
difference of likelihood scores between lncRNAs and controls (Figure S16).
Experimental validation of lncRNA candidates
We validated poly(A)+ and poly(A)- lncRNAs by RT-PCR. Poly(A)+ RNAs,
poly(A)- RNAs and total RNAs were obtained following the same procedure as
sequencing library construction. cDNAs were synthesized from total RNAs,
both of poly(A)+ RNAs and poly(A)- RNAs using the Superscript III Reverse
transcription system (Invitrogen) and Random Hexamer primers. We designed
specific primers to amplify selected lncRNAs in these cDNA libraries with
RT-PCR, using the housekeeping gene ACT2 as positive control.
We validated lncRNAs/coding genes that response to stress conditions
using qRT-PCR. The qRT-PCR was performed with Roche LightCycler®480 II
system using One Step SYBR® PrimeScript™ RT-PCR Kit (Perfect Real Time)
(TaKaRa). Expression values were normalized by ACT2.
To validate the pif4 and pifq mutants, we performed RT-PCR to test the
mutated gene. We did the DNA digestion and reverse transcription for six
samples, including Col-0, pif4 mutant and pifq mutant at normal condition and
high light condition. For the high light treatment, we used the previous reported
method(Filichkin et al., 2010). We amplified PIF4 in the pif4 mutant, and PIF1,
PIF3, PIF4 and PIF5 in the pifq mutant. Then we did qRT-PCR to validate the
differential expression for lncRNAs and PIF regulated marker genes in the six
samples, the same strategy was applied as above.
All the primers used in this study are listed in Table S5.
References
Bailey, T.L., Boden, M., Buske, F.A., Frith, M., Grant, C.E., Clementi, L.,
Ren, J., Li, W.W. and Noble, W.S. (2009) MEME SUITE: tools for motif
discovery and searching. Nucleic Acids Res, 37, W202-208.
Becker, C., Hagmann, J., Muller, J., Koenig, D., Stegle, O., Borgwardt, K.
and Weigel, D. (2011) Spontaneous epigenetic variation in the
Arabidopsis thaliana methylome. Nature, 480, 245-249.
Bonnet, E., Wuyts, J., Rouze, P. and Van de Peer, Y. (2004) Evidence that
microRNA precursors, unlike other non-coding RNAs, have lower
folding free energies than random sequences. Bioinformatics, 20,
2911-2917.
Brown, J.W., Echeverria, M. and Qu, L.H. (2003) Plant snoRNAs: functional
evolution and new modes of gene expression. Trends Plant Sci, 8,
42-49.
Burge, S.W., Daub, J., Eberhardt, R., Tate, J., Barquist, L., Nawrocki, E.P.,
Eddy, S.R., Gardner, P.P. and Bateman, A. (2013) Rfam 11.0: 10
years of RNA families. Nucleic Acids Res, 41, D226-232.
Cabili, M.N., Trapnell, C., Goff, L., Koziol, M., Tazon-Vega, B., Regev, A.
and Rinn, J.L. (2011) Integrative annotation of human large intergenic
noncoding RNAs reveals global properties and specific subclasses.
Genes Dev, 25, 1915-1927.
Charron, J.B., He, H., Elling, A.A. and Deng, X.W. (2009) Dynamic
landscapes of four histone modifications during deetiolation in
Arabidopsis. Plant Cell, 21, 3732-3748.
Chuang, T.J., Chen, F.C. and Chen, Y.Z. (2012) Position-dependent
correlations between DNA methylation and the evolutionary rates of
mammalian coding exons. Proc Natl Acad Sci U S A, 109,
15841-15846.
Du, Z., Zhou, X., Ling, Y., Zhang, Z. and Su, Z. (2010) agriGO: a GO analysis
toolkit for the agricultural community. Nucleic Acids Res, 38, W64-70.
Filichkin, S.A., Priest, H.D., Givan, S.A., Shen, R., Bryant, D.W., Fox, S.E.,
Wong, W.K. and Mockler, T.C. (2010) Genome-wide mapping of
alternative splicing in Arabidopsis thaliana. Genome Res, 20, 45-58.
German, M.A., Pillay, M., Jeong, D.H., Hetawal, A., Luo, S., Janardhanan,
P., Kannan, V., Rymarquis, L.A., Nobuta, K., German, R. et al. (2008)
Global identification of microRNA-target RNA pairs by parallel analysis
of RNA ends. Nat Biotechnol, 26, 941-946.
Gerstein, M.B., Lu, Z.J., Van Nostrand, E.L., Cheng, C., Arshinoff, B.I., Liu,
T., Yip, K.Y., Robilotto, R., Rechtsteiner, A., Ikegami, K. et al. (2010)
Integrative analysis of the Caenorhabditis elegans genome by the
modENCODE project. Science, 330, 1775-1787.
Gregory, B.D., O'Malley, R.C., Lister, R., Urich, M.A., Tonti-Filippini, J.,
Chen, H., Millar, A.H. and Ecker, J.R. (2008) A link between RNA
metabolism and silencing affecting Arabidopsis development. Dev Cell,
14, 854-866.
Gruber, A.R., Findeiss, S., Washietl, S., Hofacker, I.L. and Stadler, P.F.
(2010) Rnaz 2.0: improved noncoding RNA detection. Pac Symp
Biocomput, 69-79.
Juhling, F., Morl, M., Hartmann, R.K., Sprinzl, M., Stadler, P.F. and Putz, J.
(2009) tRNAdb 2009: compilation of tRNA sequences and tRNA genes.
Nucleic Acids Res, 37, D159-162.
Kong, L., Zhang, Y., Ye, Z.Q., Liu, X.Q., Zhao, S.Q., Wei, L. and Gao, G.
(2007) CPC: assess the protein-coding potential of transcripts using
sequence features and support vector machine. Nucleic Acids Res, 35,
W345-349.
Kozomara, A. and Griffiths-Jones, S. (2011) miRBase: integrating microRNA
annotation and deep-sequencing data. Nucleic Acids Res, 39,
D152-157.
Lamesch, P., Berardini, T.Z., Li, D., Swarbreck, D., Wilks, C., Sasidharan,
R., Muller, R., Dreher, K., Alexander, D.L., Garcia-Hernandez, M. et
al. (2012) The Arabidopsis Information Resource (TAIR): improved
gene annotation and new tools. Nucleic Acids Res, 40, D1202-1210.
Laubinger, S., Zeller, G., Henz, S.R., Sachsenberg, T., Widmer, C.K.,
Naouar, N., Vuylsteke, M., Scholkopf, B., Ratsch, G. and Weigel, D.
(2008) At-TAX: a whole genome tiling array resource for developmental
expression analysis and transcript identification in Arabidopsis thaliana.
Genome Biol, 9, R112.
Levin, J.Z., Yassour, M., Adiconis, X., Nusbaum, C., Thompson, D.A.,
Friedman, N., Gnirke, A. and Regev, A. (2010) Comprehensive
comparative analysis of strand-specific RNA sequencing methods. Nat
Methods, 7, 709-715.
Liao, Q., Liu, C., Yuan, X., Kang, S., Miao, R., Xiao, H., Zhao, G., Luo, H.,
Bu, D., Zhao, H. et al. (2011) Large-scale prediction of long non-coding
RNA functions in a coding-non-coding gene co-expression network.
Nucleic Acids Res, 39, 3864-3878.
Lister, R., O'Malley, R.C., Tonti-Filippini, J., Gregory, B.D., Berry, C.C.,
Millar, A.H. and Ecker, J.R. (2008) Highly integrated single-base
resolution maps of the epigenome in Arabidopsis. Cell, 133, 523-536.
Liu, J., Jung, C., Xu, J., Wang, H., Deng, S., Bernad, L., Arenas-Huertero,
C. and Chua, N.H. (2012) Genome-Wide Analysis Uncovers Regulation
of Long Intergenic Noncoding RNAs in Arabidopsis. Plant Cell.
Lu, Z.J., Yip, K.Y., Wang, G., Shou, C., Hillier, L.W., Khurana, E., Agarwal,
A., Auerbach, R., Rozowsky, J., Cheng, C. et al. (2011) Prediction
and characterization of noncoding RNAs in C. elegans by integrating
conservation, secondary structure, and high-throughput sequencing
and array data. Genome Res, 21, 276-285.
Luo, C., Sidote, D.J., Zhang, Y., Kerstetter, R.A., Michael, T.P. and Lam, E.
(2012) Integrative analysis of chromatin states in Arabidopsis identified
potential regulatory mechanisms for Natural Antisense Transcript
production. Plant J.
Nawrocki, E.P., Kolbe, D.L. and Eddy, S.R. (2009) Infernal 1.0: inference of
RNA alignments. Bioinformatics, 25, 1335-1337.
Oh, E., Zhu, J.Y. and Wang, Z.Y. (2012) Interaction between BZR1 and PIF4
integrates brassinosteroid and environmental responses. Nat Cell Biol,
14, 802-809.
Rabani, M., Kertesz, M. and Segal, E. (2008) Computational prediction of
RNA structural motifs involved in posttranscriptional regulatory
processes. Proc Natl Acad Sci U S A, 105, 14885-14890.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M. and Kumar, S.
(2011) MEGA5: molecular evolutionary genetics analysis using
maximum likelihood, evolutionary distance, and maximum parsimony
methods. Mol Biol Evol, 28, 2731-2739.
Trapnell, C., Roberts, A., Goff, L., Pertea, G., Kim, D., Kelley, D.R.,
Pimentel, H., Salzberg, S.L., Rinn, J.L. and Pachter, L. (2012)
Differential gene and transcript expression analysis of RNA-seq
experiments with TopHat and Cufflinks. Nat Protoc, 7, 562-578.
Wang, L., Feng, Z., Wang, X. and Zhang, X. (2010) DEGseq: an R package
for identifying differentially expressed genes from RNA-seq data.
Bioinformatics, 26, 136-138.
Washietl, S., Findeiss, S., Muller, S.A., Kalkhof, S., von Bergen, M.,
Hofacker, I.L., Stadler, P.F. and Goldman, N. (2011) RNAcode: robust
discrimination of coding and noncoding regions in comparative
sequence data. Rna, 17, 578-594.
Zeller, G., Henz, S.R., Widmer, C.K., Sachsenberg, T., Ratsch, G., Weigel,
D. and Laubinger, S. (2009) Stress-induced changes in the
Arabidopsis thaliana transcriptome analyzed using whole-genome tiling
arrays. Plant J, 58, 1068-1082.
Zhou, J., Wang, X., He, K., Charron, J.B., Elling, A.A. and Deng, X.W. (2010)
Genome-wide profiling of histone H3 lysine 9 acetylation and
dimethylation in Arabidopsis reveals correlation between multiple
histone marks and gene expression. Plant Mol Biol, 72, 585-595.