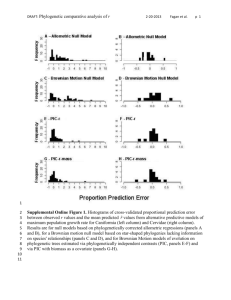
Supplemental Online Appendices
advertisement

Supplemental Online Appendix A: Development of Allometric Null Models and Comparisons to Metabolic Theory We employed allometric relationships linking body mass and r in mammals to develop allometric-null models against which to judge our PIC-based methods. Allometric models for r, which require data on body mass but not on other life history traits, have been widely used to explore the interspecific relationships in r for decades (e.g., Cole 1954, Blueweiss et al. 1978, Robinson and Redford 1986) and also feature prominently in metabolic scaling theory (e.g., Savage et al. 2004). Historically, allometric regression models did not attempt to account for phylogenetic non-independence among species in the datasets. However, phylogenetically based statistical approaches may be used in allometric analyses to incorporate covariation due to shared evolutionary history among species (Duncan et al. 2007, Fagan et al. 2010). This joint approach is most appropriate when building allometric models from existing databases which may be biased in their taxonomic representation (Fagan et al. 2010). To build our allometric null models, we used phylogenetically corrected least squares regression to account for correlated errors due to phylogenetic relatedness (Ives et al. 2007). Note that while our allometric-null models include phylogenetic information for the sample of species in the original analysis, they do not incorporate the phylogenetic position of species for which predicted values are sought. In other words, the allometric null regression models represent static mappings between female body size and r for the suite of species under consideration whereas the PIC models customize predictions for the target species based additionally on their shared evolutionary history with the rest of the clade (Garland & Ives 2000). According to metabolic theory, population growth rates, 𝑟, can be estimated based on scaling relationships with other life-history parameters, most commonly species mass, 𝑀, and age of first reproduction, 𝛽. In this appendix, we test how our PIC-r model based on comparative phylogenetics relates to metabolic theory. First, we consider whether or not our model supports the metabolic theory scaling relationships: 𝑟 ∝ 𝛽 −1 and 𝑟 ∝ 𝑀−0.25 (Savage et al. 2004, Duncan et al. 2007). To do this, we consider linear fits of 𝑙𝑜𝑔(𝑟) vs. 𝑙𝑜𝑔(𝛽) and 𝑙𝑜𝑔(𝑟) vs. 𝑙𝑜𝑔(𝑀) where 𝑟 for each species is found from equations (2-3) in the main text. Figure A.1 shows a scatterplot of 𝑙𝑜𝑔(𝑟) vs. 𝑙𝑜𝑔(𝛽) while Table A.1 defines regression lines for 𝑙𝑜𝑔(𝑟) vs. 𝑙𝑜𝑔(𝛽) assuming four alternative regression models (Fagan et al. 2010): (i) an ordinary least squares (OLS) regression, (ii) a phylogenetically corrected ordinary least squares (pgOLS) regression, (iii) a reduced major axis (RMA) regression and (iv) a phylogenetically corrected reduced major axis (pgRMA) regression. Table A.1 Allometric scaling exponents for 𝑟 vs. 𝛽 Caniformia exponent, (95% CI) Cervidae exponent, (95% CI) OLS −1.15(−1.25, −1.05) −0.42(−0.38, −0.66) pgOLS −0.95(−1.16, −0.74) −0.20(−0.50, −0.10) RMA −1.22(−1.32, −1.12) −0.58(−0.36, −0.86) pgRMA −1.25(−1.31, −1.19) −0.61(−0.99, −0.37) In keeping with metabolic theory, we see that, for both Caniformia and Cervidae, there is a negative linear relationship between 𝑙𝑜𝑔(𝑟) and 𝑙𝑜𝑔(𝛽). While the theoretically predicted -1 exponent from metabolic theory only falls within the 95% CI for the pgOLS regression of the Caniformia dataset, the predicted exponents for Caniformia are within 25% of the theoretical value. Results for Cervidae are somewhat more ambiguous; however, given the small size of the dataset (N = 15, smaller than is recommended for this type of allometric analysis [Wharton et al. 2006]) this is not surprising. Overall, then, we conclude that our data and our definition of 𝑟 in terms of life history parameters (equations (2-3) in the main text) give rise to a scaling law that shows the expected negative linear relationship between 𝑙𝑜𝑔(𝑟) and 𝑙𝑜𝑔(𝛽) with an exponent approximating the theoretically predicted 𝑟 ∝ 𝛽 −1 result. Figure A.1 Log transformed population growth rate, 𝑙𝑜𝑔(𝑟), as a function of log transformed age of first reproduction 𝑙𝑜𝑔(𝛽) for (a) the Caniformia group: Ailuridae (closed circles), Canidae (open circles), Mephitidae (closed triangles), Mustelidae (open triangles), Otariidae (closed diamonds), Phocidae (open diamonds), Procyonidae (closed squares), and Ursidae (open squares) and (b) the Cervidae group (closed circles). Both panels show linear fits for an ordinary least squares (OLS) regression (red, dashed), a phylogenetically corrected ordinary least squares (pgOLS) regression (red, solid), a reduced major axis (RMA) regression (black, dashed), and a phylogenetically corrected reduced major axis (pgRMA) regression (black, solid). Figure A.2 shows a scatterplot of 𝑙𝑜𝑔(𝑟) vs. 𝑙𝑜𝑔(𝑀) while Table A.2 defines regression lines for 𝑙𝑜𝑔(𝑟) vs. 𝑙𝑜𝑔(𝑀) assuming (i) OLS regression, (ii) pgOLS regression, (iii) RMA regression and (iv) pgRMA regression. Again, from Figure A.2, we see that there is a negative linear relationship between 𝑙𝑜𝑔(𝑟) and 𝑙𝑜𝑔(𝑀) (although the relationship is weaker than was the case for 𝑙𝑜𝑔(𝑟) and 𝑙𝑜𝑔(𝛽), as indicated by the lower R2 values for the two OLS fits in Figure A.2.a as compared to Figure A.1.a). While Table A.2 suggests wide variation in the predicted scaling exponent, the theoretical value of -0.25 falls within three of the eight 95% CIs. In addition, our data are consistent with other empirical estimates of scaling exponents. For example, Duncan et al. (2007) reported 𝑙𝑜𝑔(𝑟) vs. log(𝑀) scaling exponents of -0.35 for the Carnivora (parent phylogenetic order to the Caniformia) and -0.2 for the Artiodactyla (parent phylogenetic order to the Cervidae). -0.35 falls within the 95% CI for the OLS fit for Caniformia, while -0.2 falls within three of the four 95% CIs for the Cervidae group. Thus we conclude that our dataset is at least qualitatively consistent with metabolic theory to the extent that there is a negative linear relationship between 𝑙𝑜𝑔(𝑟) and 𝑙𝑜𝑔(𝑀). Scaling exponents approximate both theoretical predictions and results from other empirical studies based on metabolic theory. This evidence for allometric relationships between r and M, coupled with historical emphasis on such relationships (Cole 1954, Blueweiss et al. 1978, Hennemann 1984, Schmitz and Lavigne 1984, Robinson and Redford 1986, Ross 1992), justifies our use of allometric null models as benchmarks against which to judge the performance of the models based on phylogenetically independent contrasts (PIC). Table A.2 Predicted scaling relationships for 𝑟 vs. 𝑀 Caniformia exponent (95% CI) Cervidae exponent (95% CI) OLS −0.35(−0.42, −0.28) −0.10(−0.26, 0.05) pgOLS −0.10(−0.24, 0.04) 0.005(−0.14, 0.15) RMA −0.46(−0.54, −0.39) −0.28(−0.47, −0.16) pgRMA −0.49(−0.55, −0.44) −0.27, (−0.95, −0.08) Next, we ask how predicted 𝑟 values based on scaling laws from metabolic theory compare to predicted 𝑟 values from our PIC-r model. Figure A.3 shows the magnitude of the absolute prediction error, |𝑟̂ − 𝑟| as a function of 𝑙𝑜𝑔(𝑟) for the PIC-r model (black circles) and also for 𝑟 estimates based on pgOLS fits of 𝑙𝑜𝑔(𝑟) vs 𝑙𝑜𝑔(𝛽) (grey circles) and 𝑙𝑜𝑔(𝑟) vs 𝑙𝑜𝑔(𝑀) (open circles). These last two methods correspond to using the (empirically fitted) scaling relationships from metabolic theory to estimate 𝑟, with the latter being the allometric null model from the main text. Figure A.2 Log transformed population growth rate, 𝑙𝑜𝑔(𝑟), as a function of log transformed species mass 𝑙𝑜𝑔(𝑀) for (a) the Caniformia group: Ailuridae (closed circles), Canidae (open circles), Mephitidae (closed triangles), Mustelidae (open triangles), Otariidae (closed diamonds), Phocidae (open diamonds), Procyonidae (closed squares), and Ursidae (open squares). (b) the Cervidae group (closed circles). Both panels show linear fits for an ordinary least squares (OLS) regression (grey, dashed), a phylogenetically corrected ordinary least squares (pgOLS) regression (grey, solid), a reduced major axis (RMA) regression (black, dashed), and a phylogenetically corrected reduced major axis (pgRMA) regression (black, solid). Figure A.3 Magnitude of the absolute prediction error |𝑟̂ − 𝑟|, as a function of log transformed population growth rate 𝑙𝑜𝑔(𝑟) for (a) the Caniformia group and (b) the Cervidae group. Errors for the PIC-r mass model are shown in black, errors based on a pgOLS of 𝑙𝑜𝑔(𝑟) vs. 𝑙𝑜𝑔(𝛽) are shown in grey and errors based on a pgOLS of 𝑙𝑜𝑔(𝑟) vs. 𝑙𝑜𝑔(𝑀), also the allometric null model in the main text, are shown in red. From Figure A.3 it is clear that our PIC-r model performs better than the allometric null model based on pgOLS of 𝑙𝑜𝑔(𝑟) vs. 𝑙𝑜𝑔(𝑀). PIC-r offers an improvement over the model based on pgOLS of 𝑙𝑜𝑔(𝑟) vs. 𝑙𝑜𝑔(𝛽) for ~50% of species. Supplemental Online Appendix B: PIC-r Prediction Errors To understand the limitations of and biases in the PIC-r approach, we consider trends in the absolute PIC-r prediction error, (𝑟̂ − 𝑟), as a function of life-history traits. Table B.1 shows the slopes and corresponding p-values obtained from pGLS regressions of (𝑟̂ − 𝑟) against: species mass (𝑀), average interval between litters (∆), maximum number of female offspring per litter (𝑚), minimum age of first reproduction (𝛽), average mortality rate (𝜇), and maximum population growth rate (𝑟). Slopes shown in bold are significantly non-zero at the 95% confidence level. Table B.1 pGLS regressions of absolute prediction errors (𝑟̂ − 𝑟) vs. life-history parameters LifeHistory Parameter Caniformia Cervidae slope p-value slope p-value 𝑀 0.0003 0.88 5.0×10-5 0.85 ∆ 0.29 0.088 0.24 0.046 𝑚 -0.42 4.1×10-8 -0.23 0.037 𝛽 0.13 0.048 0.032 0.52 𝜇 -1.71 0.030 -0.37 0.78 𝑟 -1.02 0 -1.09 4.24×10-8 Based on results in Table B.1, it is clear that absolute prediction error correlates with a number of life-history parameters. For example, large litters are associated with underestimation of 𝑟 while small litters are associated with overestimation of 𝑟. Similar trends can be seen for mortality rate and age of first reproduction in Caniformia, and the interval between litters in Cervidae. It is tempting to assume that these dependencies arise as a result of correlations between life-history traits and key aspects of evolution (e.g. rate), and thus reflect inaccuracies in estimation of the underlying phylogenetic tree (e.g. branch length). However, there is a much simpler explanation: 𝑟 scales with each of the life-history parameters shown in Table A.1. Moreover, because PIC-r predicts a focal species’ 𝑟 based on the weighted average of 𝑟 values for other species in the tree, absolute prediction errors naturally exhibit a negative dependence on 𝑟 itself (i.e. predictions for species with large 𝑟 tend to be underestimates while predictions for species with small 𝑟 tend to be overestimates). Absolute prediction errors thus scale with lifehistory parameters by virtue of 𝑟 scaling with life-history parameters and absolute prediction errors scaling with 𝑟. To see the argument above, consider a null model wherein estimates of a species’ 𝑟 are based on non-phylogenetically corrected averages of the 𝑟 values of all other species: 𝑟̂𝑖𝑛𝑢𝑙𝑙 = ∑𝑛≠𝑖 𝑟𝑛 𝑁−1 (B.1) In equation (B.1), 𝑟̂𝑖𝑛𝑢𝑙𝑙 is the estimate of 𝑟 for species 𝑖, 𝑟𝑛 is the known value of 𝑟 for species 𝑛, and there are 𝑁 species in the dataset (including the focal species 𝑖). For large 𝑁, the right hand side of equation (B.1) will be very nearly constant for all 𝑖, thus the absolute prediction error, (𝑟̂𝑖𝑛𝑢𝑙𝑙 − 𝑟), will be close to a straight line with a slope of -1. This simple example illustrates why averaging tends to give high estimates for species with low 𝑟 and low estimates for species with high 𝑟. In Figure B.1, we show absolute prediction error as a function of maximum population growth rate for the PIC-r model of the Caniformia. We also show an OLS regression of absolute prediction error against maximum population growth rate for the null model in equation (B.1) (black dashed line). To the extent that PIC-r prediction errors fall between the black dashed line and its reflection about the x-axis (red dashed line), phylogenetic correction improves estimates as compared to the null model. However, despite this improvement, the negative trend in absolute prediction error with maximum population growth rate is retained, explaining error dependencies on other life-history traits as well. Figure B.1 Absolute prediction error (𝑟̂ − 𝑟) as a function of maximum population growth rate 𝑟. Prediction errors from the PIC-r model of the Caniformia group are shown as: Ailuridae (closed circles), Canidae (open circles), Mephitidae (closed triangles), Mustelidae (open triangles), Otariidae (closed diamonds), Phocidae (open diamonds), Procyonidae (closed squares), and Ursidae (open squares). Prediction errors from the null model in equation (B.1) are shown as an OLS regression (black dashed line). The grey dashed line is the reflection of the black dashed line through the x-axis. PIC-r prediction errors falling between the red and black dashed lines indicate an improvement in PIC-r as compared to the basic null model in equation (B.1). Interestingly, the only life-history parameter that does not seem to have a strong effect on PIC-r prediction error is species mass, 𝑀. Indeed, for both Caniformia and Cervidae, slope estimates from pGLS regressions of prediction error against mass are very nearly zero and not significantly different from zero. Figure B.2 shows scatterplots of PIC-r prediction error as a function of mass. Again, the apparent lack of correlation is obvious to the eye: over- and under- estimates of 𝑟 appear equally likely regardless of species size. We suggest that this apparent lack of correlation between prediction error and mass is at the root of the improved performance of the simple PIC-r model relative to the more complex PIC-r-mass model. An alternative possibility, wherein noise in the 𝑙𝑜𝑔(𝑟) vs 𝑙𝑜𝑔(𝑀) relationship is responsible for the lack of improvement when using the PIC-r-mass model, is not supported by the data. Outlier species in the allometric regression are neither more nor less likely to be better estimated via the PIC-r-mass model than by the PIC-r model (Fig. B3). Figure B.2 Absolute prediction error (𝑟̂ − 𝑟) as a function of species mass 𝑀. (a) Prediction errors from the PIC-r model of the Caniformia group are shown as: Ailuridae (closed circles), Canidae (open circles), Mephitidae (closed triangles), Mustelidae (open triangles), Otariidae (closed diamonds), Phocidae (open diamonds), Procyonidae (closed squares), and Ursidae (open squares). (b) Prediction errors from the PIC-r model of the Cervidae group are shown as closed circles. Figure B.3 Log(r) vs. log(M) for Caniformia and Cervidae. Species for which PIC-r-mass performs better are denoted by green circles. Species for which PIC-r performs better are denoted by red circles. The black dotted lines indicate phylogenetically corrected RMA fits. Supplemental Online Appendix C: Raw Data for Caniformia and Cervidae Supplemental Table C.1 Caniformia life-history parameters Species r M (kg) (yr) (yr) µ (yr-1) m (#) tip length Ailuropoda melanoleuca Ailurus fulgens 0.139544 0.387016 120 4.33 1.43 1.00 6.44 1.63 0.05 0.15 0.77 1.01 0.453258 0.875254 Amblonyx cinereus 0.574116 3 0.50 1.08 0.18 0.71 0.192695 Arctocephalus australis 0.195119 45 1.00 3.00 0.07 0.50 0.012764 Arctocephalus forsteri 0.141511 55 1.00 5.00 0.06 0.50 0.02031 Arctocephalus galapagoensis 0.086798 27 2.00 3.50 0.08 0.50 0.012764 Arctocephalus gazella 0.200314 40.5 1.00 2.83 0.07 0.50 0.006099 Arctocephalus pusillus 0.187227 77.67 1.00 3.54 0.05 0.50 0.099173 Arctocephalus tropicalis 0.16608 50 1.00 4.00 0.06 0.50 0.006099 Bassariscus astutus 0.644558 0.976 1.00 0.89 0.27 1.35 0.202692 Callorhinus ursinus 0.148494 42.4 1.12 4.08 0.06 0.50 0.208686 Canis adustus 1.81319 8.25 0.50 0.64 0.12 2.15 0.124854 Canis latrans 1.097966 11.8 1.00 1.14 0.10 2.75 0.092248 Canis lupus 0.710635 27 1.22 1.82 0.07 2.57 0.092248 Canis mesomelas 1.0542 9.75 1.00 0.90 0.11 1.98 0.26084 Cerdocyon thous 1.51811 6.5 0.53 0.76 0.13 2.05 0.06524 Cystophora cristata 0.223528 212 1.00 2.99 0.04 0.50 0.166204 Eira barbara 1.4694 4.5 0.50 0.50 0.15 1.25 0.262033 Enhydra lutris 0.165412 21.8 1.15 2.69 0.09 0.50 0.28868 Gulo gulo 0.302902 16.3 2.22 2.23 0.09 1.42 0.331999 Halichoerus grypus 0.174065 182 1.08 4.21 0.04 0.50 0.046263 Ictonyx striatus 0.549191 0.765 1.00 0.77 0.29 1.09 0.165972 Lobodon carcinophagus 0.190796 238 1.00 4.01 0.04 0.50 0.229426 Lontra canadensis 0.351403 4.6 1.06 2.50 0.14 1.36 0.336471 Lutra lutra 0.576544 6.75 1.00 1.00 0.13 1.03 0.192695 Martes americana 0.367861 0.607 1.00 1.47 0.32 1.37 0.300865 Martes pennanti 0.536204 2.6 1.00 1.44 0.19 1.46 0.262033 Meles meles 0.687938 13 1.33 1.10 0.10 1.56 0.498783 Mephitis macroura 1.12746 1.2 1.00 0.80 0.25 2.25 0.08313 Mephitis mephitis 1.19265 2 0.83 0.93 0.21 2.55 0.08313 Monachus schauinslandi 0.127559 173 1.44 5.00 0.04 0.50 0.324987 Mustela erminea 4.10122 0.11 1.00 0.26 0.60 3.39 0.153847 Mustela eversmannii 1.69721 1.35 1.00 0.88 0.24 4.71 0.014018 Mustela frenata 1.813366 0.151 1.00 0.53 0.48 3.05 0.145605 Mustela nigripes 0.673729 0.703 1.00 1.00 0.30 1.65 0.028501 Mustela nivalis 2.76656 0.0498 0.60 0.31 0.81 2.69 0.123539 Mustela putorius 1.599601 0.65 0.79 0.83 0.31 3.84 0.014018 Mustela vison 1.26146 0.9 1.00 0.72 0.28 2.38 0.145605 Nasua narica 0.49902 4.6 1.00 2.00 0.15 1.75 0.586311 Neophoca cinerea 0.161991 79.6 1.45 3.00 0.05 0.50 0.081416 Nyctereutes procyonoides 1.55555 4.23 1.00 1.02 0.16 4.73 0.199755 Ommatophoca rossii 0.202926 188 1.00 3.46 0.04 0.50 0.229426 Otaria byronia 0.192369 140 1.00 3.69 0.04 0.50 0.106778 Otocyon megalotis 1.18362 4.15 0.50 1.00 0.16 1.87 0.304675 Phoca fasciata 0.233474 82.5 1.00 2.45 0.05 0.50 0.124444 Phoca groenlandica 0.177911 128.8 1.00 4.15 0.04 0.50 0.124444 Phoca hispida 0.111449 69.7 1.25 6.16 0.05 0.50 0.036628 Phoca sibirica 0.155588 81.7 1.00 4.82 0.05 0.51 0.036628 Phoca vitulina 0.181563 101 1.00 3.88 0.05 0.50 0.063008 Phocarctos hookeri 0.201567 183 1.00 3.50 0.04 0.50 0.087359 Poecilogale albinucha 0.30648 0.2575 0.80 1.20 0.44 1.10 0.165972 Potos flavus 0.120924 3 1.00 2.27 0.17 0.50 0.803102 Procyon lotor 0.886438 4.4 1.00 0.95 0.14 1.72 0.202692 Pseudalopex culpaeus 0.99461 13 1.00 1.00 0.10 2.00 0.06524 Pteronura brasiliensis 0.390658 24 1.00 2.00 0.08 0.97 0.38378 Speothos venaticus 1.197471 6 0.57 0.96 0.14 1.93 0.163721 Spilogale putorius 1.679876 0.512 0.82 0.51 0.36 2.31 0.273302 Taxidea taxus 0.976827 6.05 1.35 0.61 0.14 1.54 0.550177 Urocyon cinereoargenteus 1.096611 3.3 1.01 0.94 0.17 2.39 0.504537 Ursus americanus 0.197258 87.2 2.41 3.68 0.05 1.10 0.185153 Ursus arctos 0.148576 204 2.89 5.17 0.04 1.09 0.185153 Vulpes macrotis 0.89761 1.9 1.00 1.00 0.20 2.00 0.101789 Vulpes vulpes 1.15437 3.65 1.10 0.81 0.14 2.17 0.101789 Vulpes zerda 0.642439 1.2 1.00 0.82 0.25 1.23 0.258465 Zalophus californianus 0.137426 84.7 1.00 5.67 0.05 0.50 0.123212 Supplemental Figure C.1 Caniformia Phylogenetic Tree from Agnarsson et al. 2010 Supplemental Table C.2 Cervidae life-history parameters Species r M (kg) (yr) (yr) µ (yr-1) m (#) tip length Alces alces Axis axis Cervus albirostris Cervus elaphus Cervus eldii Cervus nippon Cervus timorensis Cervus unicolor Dama dama Elaphurus davidianus Mazama americana Muntiacus reevesi Odocoileus hemionus Ozotoceros bezoarticus Rangifer tarandus 0.361242 0.34528 0.271815 0.303467 0.296612 0.26731 0.204153 0.273818 0.282863 0.277142 0.318766 0.552882 0.43003 0.33528 0.399106 408 50 125 109 73 96.5 53 171 54.5 149 23 12 56.3 35 93.6 1.13 1.00 1.00 1.00 1.00 1.00 1.50 1.00 1.00 1.00 1.00 0.67 1.03 1.00 0.95 2.22 1.00 2.00 1.52 1.57 1.84 1.74 1.97 1.79 2.25 1.05 0.50 1.43 1.00 1.00 0.03 0.06 0.04 0.05 0.05 0.06 0.06 0.04 0.06 0.04 0.08 0.11 0.06 0.07 0.05 0.86 0.50 0.50 0.50 0.51 0.51 0.50 0.50 0.54 0.56 0.50 0.50 0.96 0.50 0.55 16.6 12.5 7.5 7.4 7.5 7.5 7.6 7.4 12.2 7.6 4 17 4.1 4 11.5 Supplemental Figure C.2 Cervidae Phylogenetic Tree from Bininda-Emonds et al. 2007