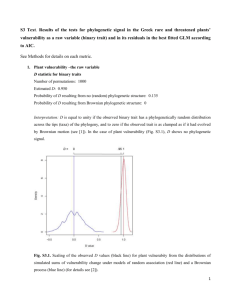
Supplemental Online Figures - Proceedings of the Royal Society B
advertisement
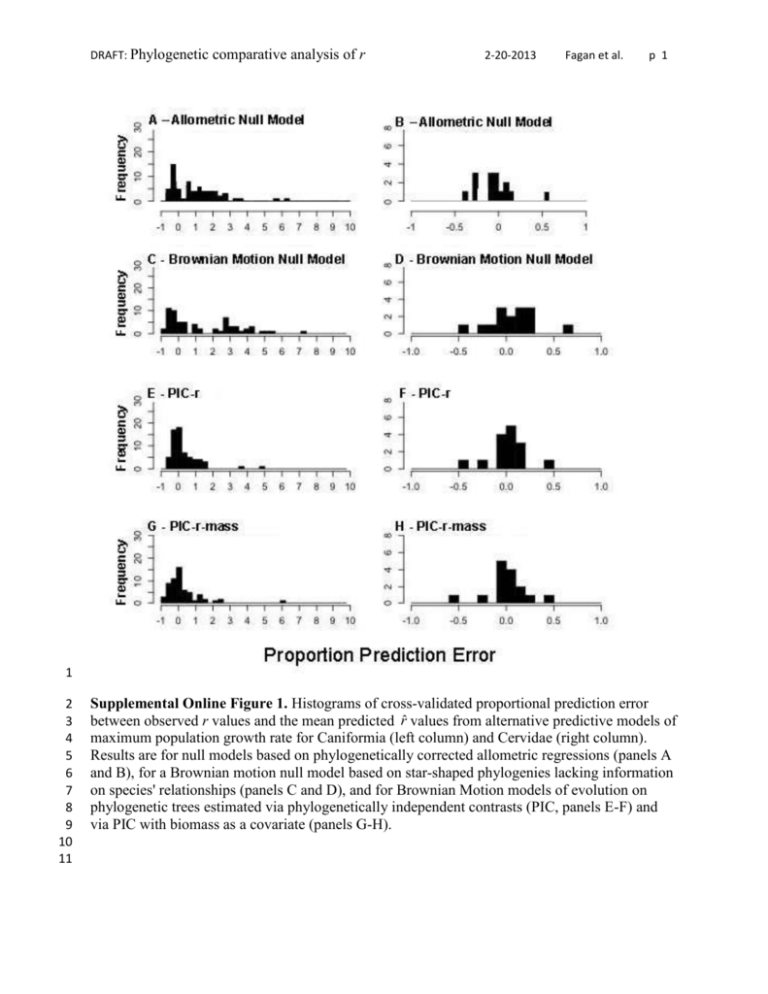
DRAFT: Phylogenetic comparative analysis of r 2-20-2013 Fagan et al. p 1 1 2 3 4 5 6 7 8 9 10 11 Supplemental Online Figure 1. Histograms of cross-validated proportional prediction error between observed r values and the mean predicted r̂ values from alternative predictive models of maximum population growth rate for Caniformia (left column) and Cervidae (right column). Results are for null models based on phylogenetically corrected allometric regressions (panels A and B), for a Brownian motion null model based on star-shaped phylogenies lacking information on species' relationships (panels C and D), and for Brownian Motion models of evolution on phylogenetic trees estimated via phylogenetically independent contrasts (PIC, panels E-F) and via PIC with biomass as a covariate (panels G-H). DRAFT: Phylogenetic comparative analysis of r 2-20-2013 Fagan et al. p 2 12 13 14 15 16 17 18 19 20 21 Supplemental Online Figure 2. Relationship between predicted r as estimated by the allometric null model, and predicted r as estimated by the PIC-r model for (a) Caniformia and (b) Cervidae. Points that fall along the dashed 1:1 line represent identical predicted values of r from both models. Solid lines represent an ordinary least squares fit (Caniformia: y = 1.14x - 0.17, R2 = 0.62, Cervidae: y = 0.26x + 0.24, R2 = 0.09) of the relationship between predictions from the Allometric Null LS model and predictions from the PIC-r model. In (a) individual families are: Ailuridae (closed circles), Canidae (open circles), Mephitidae (closed triangles), Mustelidae (open triangles), Otariidae (closed diamonds), Phocidae (open diamonds), Procyonidae (closed squares), and Ursidae (open squares). DRAFT: Phylogenetic comparative analysis of r 2-20-2013 Fagan et al. p 3 Supplemental Online Figure 3. Output from the allometric null and Brownian motion null models for Caniformia providing a comparison of observed r values (filled circles) and r̂ values predicted by cross-validation (open circles). Error bars on each model result give 1 SE (inner pair) and 95% PI (outer pair) of the null model prediction. DRAFT: Phylogenetic comparative analysis of r 2-20-2013 Fagan et al. p 4 Supplemental Online Figure 4. Output from the allometric null and Brownian motion null models for Cervidae providing a comparison of observed r values (filled circles) and r̂ values predicted by cross-validation (open circles). Error bars on each model result give 1 SE (inner pair) and 95% PI (outer pair) of the null model prediction. DRAFT: Phylogenetic comparative analysis of r 2-20-2013 Fagan et al. p 5 DRAFT: Phylogenetic comparative analysis of r 2-20-2013 Fagan et al. p 6 Supplemental Online Figure 5. Comparison of observed r values (filled circles) and r̂ values predicted by leave-one-out crossvalidation (open circles) for the Caniformia (Panel A) and the Cervidae (Panel B) using the predictions for the PIC-r (first and orange) and the PIC-r-mass (second, blue). Error bars on each model result give 1 SE (inner pair) and 95% PI (outer pair). The data for PICr are plotted phylogenetically in Figure 2 in the main text. DRAFT: Phylogenetic comparative analysis of r 2-20-2013 Fagan et al. p 7 Supplemental Online Figure 6. Cross-validated proportional prediction error between observed r values and the mean predicted r̂ values from two phylogenetically structured evolutionary models of maximum population growth rate for Caniformia. This figure plots data from Figure 2 on the phylogenetic tree to illustrate that large prediction errors are not strongly clustered phylogenetically. The diameters are proportional to error magnitude (maximum = 6.11); white circles are negative values (i.e., are underestimates). DRAFT: Phylogenetic comparative analysis of r 2-20-2013 Fagan et al. p 8 Supplemental Online Figure 7. Cross-validated proportional prediction error between observed r values and the mean predicted r̂ values from two phylogenetically structured evolutionary models of maximum population growth rate for Cervidae. This figure plots data from Figure 2 on the phylogenetic tree to illustrate that large prediction errors are not strongly clustered phylogenetically. The diameters are proportional to error magnitude (maximum = 0.54); white circles are negative values (i.e., are underestimates).