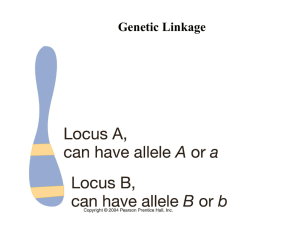
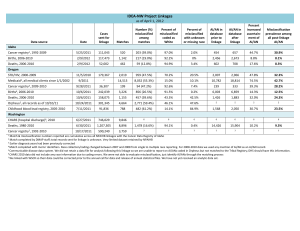
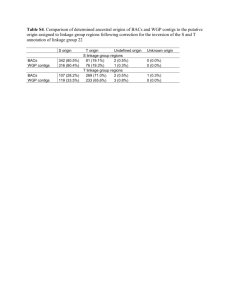
Key Points. Linkage.
advertisement

Key Points. Linkage. Why is it that sister chromatid exchange, as long as it is reciprocal, does not lead to new combinations of alleles at linked loci? If there was loss or gain of chromatin with each crossover event, do you think that crossing over would have evolved as such a potent source of genetic variation? What are the consequences (if any) – in terms of segregation and in terms of independent assortment – of the random alignment of non-homologous chromosomes on the Metaphase I “plate”? Consider the following data from a testcross in a tropical flower. Purple non-aromatic 42 Purple aromatic 8 White nonaromatic 6 White aromatic 44 Which phenotypes are non-parental? What is the distance (expressed as % recombination) between the color and aroma loci? If you converted the % recombination to cM, would the value be larger or smaller? Is recombination at meiosis a source of new alleles, a source of new combinations of alleles at linked loci, or both? How can you determine if the association of two traits in inheritance is due to linkage or pleiotropy? If you discover that a negative association between two traits is due to pleiotropy (e.g. good flavor and disease susceptibility), what option(s) do you have to develop a variety of your favorite plant that has good flavor and is disease resistant? You read that a species is 2n = 28 but when you make a linkage map you get 22 linkage groups. What are two possible explanations? Define the following terms, as they relate to linkage: Chiasma Sister chromatid Non-sister chromatid Cis/coupling Trans/repulsion Explain how two loci on the same chromosome can show independent assortment. The examples of linkage analysis have all involved doubled haploid or test cross examples. Is it possible to do linkage analysis with an F2? Why are molecular markers more popular for linkage map construction than naked eye polymorphisms? Differentiate between homology, homoeology, synteny, and orthology.