Evaluation of whole genome amplification from small cell numbers
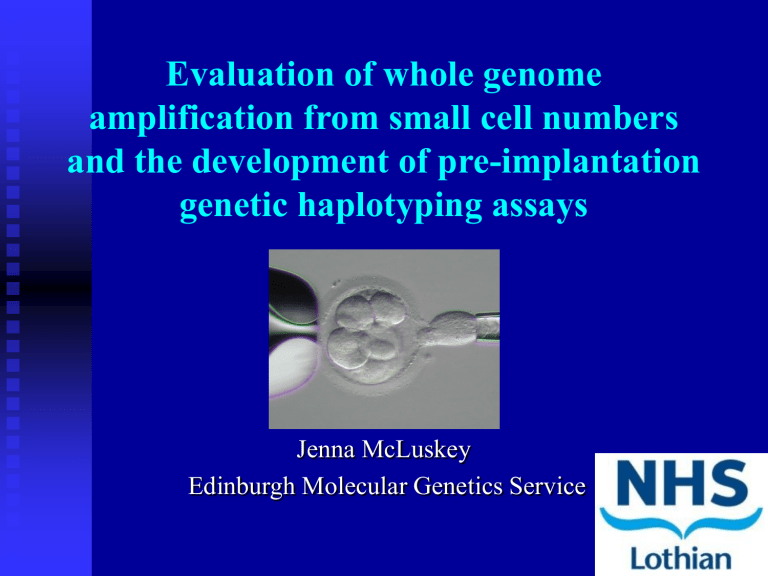
Evaluation of whole genome amplification from small cell numbers and the development of pre-implantation genetic haplotyping assays
Jenna McLuskey
Edinburgh Molecular Genetics Service
Preimplantation Genetic Diagnosis (PGD)
Hormonal stimulation of the ovaries to produce mature oocytes.
Intracytoplasmic sperm injection (ICSI) or in vitro fertilisation (IVF).
Embryo Biopsy
Genetic analysis of one or two cells
- PCR or FISH.
IVF
Embryo Development following fertilisation (day 0-2)
ICSI
Fertilised egg 2 cell embryo 4 cell embryo
Cleavage stage biopsy
Project Aims
Whole genome amplification: small cell numbers
- Buccal cells: 1 / 2 /5 / multiple cells
- (Blastomeres:1 / 2 /5 / multiple cells ?)
Theoretical microsatellite marker evaluation, validation & incorporation into multiplex assays.
Marker segregation analysis.
Application of multiplex assays to WGA products.
Schematic of buccal cell collection, rinsing and lysis
Whole Genome Amplification (WGA)
Produces large quantities of DNA from small templates.
Overcomes problems with single cell lysates.
Successful PCR amplification, following
WGA for 5/5 single lymphocytes and 10/11 single blastomeres .
Handyside et al (2004) Isothermal whole genome amplification from single and small numbers of cells: a new era for PGD of inherited diseases. Molecular Reproduction 10;
767-772
Whole Genome Amplification:
Multiple Displacement Amplification
(MDA)
A. Mamone, 2003, Amersham Biosciences, Piscataway, NJ, USA
MDA products electrophoresed on a 2% agarose gel
L 1 2 3 4 5 6 7 8 9 10 L L 1 2 3 4 5 6 7 8 9 L
L 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 L
L 1 2 3 4 5 6 7 8 9 10 11 12 L
Validation of WGA DNA products
Amplification products assessed using
QF-PCR assay for the detection of common aneuploidies.
12 tetra nucleotide repeat markers for chromosomes 13, 18 and 21.
PCR products amplified from WGA DNA vs DNA extracted from blood lymphocytes.
blood lymphocytes five buccal cells
D21S1437 D21S11 D13S628 D13S634 D18S535 blood lymphocytes five buccal cells
D18S1002 D18S391 D13S742 D18S386 D13S305 blood lymphocytes five buccal cells
IFNAR D211411
blood lymphocytes five buccal cells
D21S1437 D21S11 D13S628 D13S634 D18S535 blood lymphocytes five buccal cells
D18S1002 D18S391 D13S742 D18S386 D13S305 blood lymphocytes five buccal cells
IFNAR D211411
blood lymphocytes five buccal cells
D21S1437 D21S11 D13S628 D13S634 D18S535 blood lymphocytes five buccal cells
D18S1002 D18S391 D13S742 D18S386 D13S305 blood lymphocytes five buccal cells
IFNAR D211411
blood lymphocytes five buccal cells
D21S1437 D21S11 D13S628 D13S634 D18S535 blood lymphocytes five buccal cells
D18S1002 D18S391 D13S742 D18S386 D13S305 blood lymphocytes five buccal cells
IFNAR D211411
Direct mutation testing vs haplotyping
Allele drop out (ADO) more disruptive to direct mutation test:
- False positives
- False negatives
number of markers,
chances of a result.
Preimplantation Genetic Haplotyping (PGH)
Applicable to any single gene disorder in which the causative gene has been mapped.
Microsatellite markers span disease locus.
Multiplex assays create dense haplotypes
Renwick et al – 12 closely linked microsatellite markers – 93% haplotypes constructed despite some ADO at individual loci.
Renwick et al (2006) Proof of Principle and first cases using PGH – a paradigm shift for embryo diagnosis. Reproductive Medicine 13; 758-767
Guys’ and St Thomas’ two tube PGH assay for Cystic Fibrosis (CF)
PGH for CF currently offered at Guys’ and
St Thomas’ Hospital, London.
Two tube universal tagged fluorescent multiplex.
Ten dinucleotide & 3 tetranucleotide repeat markers spanning the CFTR locus.
D7S523
5.4Mb
Guys’ and St Thomas’ two tube PGH assay for Cystic Fibrosis (CF)
D7S2554
2.7 Mb
D7S486
1.2 Mb
IVS1CA
69.4 Kb
Phe508 CFSTR1
0.3 Mb
D7S643
3.6 Mb
D7S650
3.7Mb
D72490
5.5Mb
D7S2502
1.7Mb
D7S2460
0.7Mb
IVS8CA
11.3Kb
D7S2847
1.5 Mb
D7S480
3.7Mb
D7S523
5.4Mb
Removal of two least useful markers
D7S2554
2.7 Mb
D7S486
1.2 Mb
IVS1CA
69.4 Kb
Phe508 CFSTR1
0.3 Mb
D7S643
3.6 Mb
D7S650
3.7Mb
D72490
5.5Mb
D7S2502
1.7Mb
D7S2460
0.7Mb
IVS8CA
11.3Kb
D7S2847
1.5 Mb
D7S480
3.7Mb
Selection and evaluation of theoretical polymorphic markers
1.
2.
3.
4.
5.
20 microsatellite markers selected.
Primer selection using Primer 3.
Markers evaluated individually.
Incorporate markers into assay.
Calculate Polymorphism Information Content
(PIC) & Heterozygosity (HET) scores.
PIC & HET values
Marker HET Score PIC Score
MS1 0.87
0.86
MS3
MS6
MS7
MS15
MS19
0.90
0.76
0.53
0.68
0.86
0.89
0.72
0.51
0.64
0.84
(n=192)
Addition of new markers to two tube PGH assay for Cystic Fibrosis (CF)
MS1
4.6Mb
D7S2554
2.7 Mb
MS3
0.7 Mb
D7S486
1.2 Mb
IVS1CA
69.4 Kb
Phe508 CFSTR1
0.3 Mb
MS6
2.6 Mb
D7S643
3.6 Mb
D7S650
3.7Mb
D7S2502
1.7Mb
D7S2460
0.7Mb
IVS8CA
11.3Kb
MS19
0.4Mb
D7S2847
1.5 Mb
D7S480
3.7Mb
Multiplex A
Multiplex B
Typical CF haplotypes from family analysis
Buccal cells vs lymphocytes
WGA of blastomeres
Discarded embryos.
Embryo’s biopsied.
Single cell removed and lysed.
Remainder of embryo lysed (used as comparison).
1/10
Preliminary embryo data
1/6
5/6
1/6 1/10
5/6 9/10
Conclusions
WGA from small cell numbers was successful.
Four new polymorphic markers found close to CFTR .
Markers incorporated into CF assay.
Highly informative haplotypes –universally applicable.
Assay suitable for WGA DNA from small cell numbers.
Preliminary embryo data is promising!
Acknowledgements
Pamela Renwick, Jane Trussler & Cheryl Black
(Guys’ and St Thomas’Hospital, London).
Sue Pickering (Assisted Conception Unit,
Edinburgh).
Jon Warner & Paul Westwood (Edinburgh
Molecular Genetics).
Everyone in the Edinburgh Molecular Genetics
Lab.