Lecture 1
advertisement

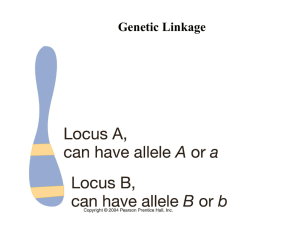
Hunting Disease Genes in the Wilds of the Genome -- I HMGP Richard A. Spritz, M.D. March 29, 2011 richard.spritz@ucdenver.edu 303-724-3107 Why Find Disease Genes? Disease Gene Identification— “Functional Cloning” vs. “Positional Cloning” Positional Cloning: Determine a Disease Gene’s Genomic Position, and then Identify the Gene Obviated by Human Genome Project Gene Mapping Technology Polymorphic DNA Markers • • • • You can only track/measure differences between people and through families Polymorphic DNA markers constitute any scoreable differences at known genomic positions Surrogates for disease mutations; some polymorphisms cause disease; most don’t Most commonly used marker types: – microsatellites – single-nucleotide polymorphisms (SNPs) – copy-number variations (CNVs) The First Goal of the HGP was to Assemble a High-Density Genome Map of Polymorphic Markers Genetic Linkage Studies •Search for regions of genome that are systematically co-inherited along with disease on passage through families •Requires families with multiple affected relatives (multiplex families) •Best at detecting genes with Mendelian effects (uncommon alleles with strong effects) •Unit of genetic linkage is LOD (“Log of the Odds) score (>3) Genetic linkage— Two loci close together on a chromosome tend not to be separated by recombination Principle of genetic linkage— Loci close by on a chromosome tend not to be separated by recombination vs. loci far apart Loci on the same chromosome Loci on different chromosomes Very close Nearby Far Apart Freq. of crossover Rare between 2 loci Some Frequent - Linkage Tight Some Absent Absent 0% 1-49% 50% Recombination 50% • Unit of genetic “distance” is centiMorgan (cM) = 1% recombination/meiosis; ~ 1 Mb “Genetic linkage analysis” Co-segregation of disease gene in “multiplex families” with alleles of polymorphic DNA “markers” (initially RFLPs) Restriction Fragment Length Polymorphism (RFLP) EcoRI Allele 1 AGAGCCTCAACTTGAATTCGTTTAGTAA Allele 2 AGAGCCTCAACTTGAATTTGTTTAGTAA Restriction enzyme EcoRI cuts at sequence 5’-GAATTC-3’ Allele 1 has an EcoRI cut site; Allele 2 does not • This RFLP is assaying a SNP “Microsatellites” (SSLPs; STRPs, SSRs) [multi-allelic; ~ 1/30,000 bp; mostly used for linkage analysis, forensics] ggctgcacacacacacacacacacacacatgctt ggctgcacacacacacacacacacacatgctt ggctgcacacacacacacacacacatgctt ggctgcacacacacacacacacatgctt ggctgcacacacacacacacatgctt Can follow “segregation” of ancestral “haplotypes” of linked marker alleles in families Recombination events prune marker haplotypes, defining “genetic interval” that must contain the disease gene •Only true for Mendelian traits •For polygenic traits, linkage can only localize disease genes within rough probability distribution of location Genetic Linkage Analysis • Statistical measure is LOD (log of odds) score LOD = Log10 Likelihood of data if loci linked at Likelihood of data if loci unlinked • Significance level: LOD >3.0 for Mendelian trait LOD >3.3 for Polygenic trait Single-Nucleotide Polymorphisms (SNPs) [bi-allelic; ~1/50-300 bp; mostly used for association analysis] SNP1 Allele 1 CCGAGATCCAGAAATCCTGAACATAA SNP1 Allele 2 CTGAGATCCAGAAATCCTGAACATAA SNP2 Allele 1 CCGAGATCCAGAAATCCTGAACATAA SNP2 Allele 2 CCGAGATCCAGAAAGCCTGAACATAA • Occurrence/allele frequencies differ in different ethic groups/populations • Can be in genes (~4,000,000) on not (~8,000,000), can result in amino acid substitutions or not • Each occurs in local context (haplotype) of surrounding SNPs (in example above, SNP2 is on background of SNP1 C allele) Haplotype Map of Human Genome International HapMAP Project •Recombination breaks macro-patterns of polymorphic genotypes on the same chromosome into haplotypes •Recombination is not truly random, so very close polymorphism genotypes carried on the same chromosome cluster into ~10-50 kb haplotype blocks in which SNP alleles are in linkage disequilibrium (marker alleles within blocks tend to be co-inherited on same piece of DNA, because recombination within blocks is uncommon); blocks smaller in African than Caucasian or Asian pops. because African pop. is more ancient •HapMap genotyped SNPs in different populations to characterize haplotype block distributions Copy-Number Variants (CNVs) [bi-allelic] Basically are common genomic deletions, hundreds to tens of thousands of nucleotides in size May be detected by LD with local SNP patterns: Allele --1---1---1----1---2----1----2----1-----1----2----1----1----1----1--Allele --2---2---2----1---1----2----2----1-----1----2----2----2----1----2--CNV Allele --1---1—[ ]--1----2--- • Tens of thousands known • Like SNPS, occurrence/allele frequencies differ in different ethic groups/populations • Individually most are rare (< 1%), collectively common • Can be in genes or not, can include genes • NOT commonly definitively causal for human disease 1000 Genomes Project, UK10K Project International projects to sequence 1000/10000 genomes from different ethnic groups • Will catalog human genetic variations (particularly SNPs) – Essential for sequence-based analysis of rare variants that may be causal for common diseases