Dna rEPLICATION - Manning's Science
advertisement

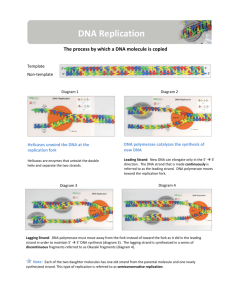
DNA REPLICATION SBI4U Ms. Manning DNA Replication Produces two identical copies of the chromosome during S phase of interphase Catalyzed by many enzymes including: DNA polymerase, helicase, ligase and primase Replication takes place at a point where the DNA double helix separates (called the replication fork) STEPS OF DNA REPLICATION 1. UNWINDING AND SEPARATION OF THE PARENT STRAND – “Initiation” -separation is initiated at one or more sites called ORIGINS OF REPLICATION -the structure of 2 unwound DNA strands is referred to as a replication fork Three enzymes work together to unwind and stabilize the double helix: A. B. C. DNA HELICASE – breaks the hydrogen bonds that connect the nitrogenous bases which allows the double helix to unwind and separate. DNA GYRASE – an enzyme that relieves the tension produced by unwinding of DNA – is a type II topoisomerase. SINGLE STRANDED BINDING PROTEINS (SSBs) – work to keep separated strands of DNA apart gyrase 2. DNA SYNTHESIS (“Elongation” Replication begins in 2 directions from the origins as a region of DNA is unwound. Replication proceeds towards the direction of the replication fork on one strand, and away from the fork on the other. In eukaryotes, more than one replication fork may exist on a DNA molecule. A replication bubble forms when 2 replication forks are in close proximity to each other Fig 5.17 p.223 DNA Synthesis cont’d… DNA POLYMERASE III takes free nucleotides found within the cell and adds them in the 5’ 3’ direction to form the new strand The parent strand is used as a template i.e. If A is in the parent, then T is inserted into the daughter LEADING STRAND The daughter strand that grows continuously towards the replication fork as the double helix unwinds. This occurs quickly The new strand grows from 5’ 3’ LAGGING STRAND The other daughter strand that cannot grow towards the replication fork, therefore it grows in the opposite direction It is built in short segments (in the 5’ 3’ direction) away from the replication fork. This is much slower than the leading strand! Details about the lagging strand: The 3’ to 5’ parent strand is a problem for DNA polymerase since it must synthesize in the 5’ 3’ direction Short RNA primer sequences of 10-60 RNA bases are bonded to regions of the lagging strand with the purpose of initiating DNA replication PRIMASE – an enzyme that binds the RNA primers to the DNA Lagging strand cont’d… DNA polymerase builds short sequences of DNA off of the RNA primers called OKAZAKI fragments. This lagging strand…is…slow! The lagging strand takes longer to replicate than the leading strand because it involves more steps and is not continuous replication. 2b) LINKING OF NITROGENOUS BASES Nitrogen bases of nucleotides of opposite strands (parent and daughter) form new H bonds Once hydrogen bonds are formed, DNA automatically twists into a double helix 3) Termination DNA polymerase I excises the RNA primers and replaces them with the appropriate deoxyribonucleotides. DNA ligase joins the gaps in the Okazaki fragments by the creation of a phosphodiester bond. 4) QUALITY CONTROL – “proofreading” DNA POLYMERASE III & I – act as a proof-reader by checking the newly synthesized strand for any incorrectly inserted bases. If a mistake is found, polymerase acts as an EXONUCLEASE – cutting out the mis-paired base and replacing it with the correct nucleotide. Errors missed by proofreading can be corrected by one of several repair mechanisms after replication is complete. What happens at the ends of chromosomes? Telomeres Shrinking telomeres and aging