The Cell Nucleus (The Living Cell) UNFINISHED
Learning Outcomes At the end of this session students should be able to: Describe the basic structure and functions of the mammalian cell nucleus Describe the structure of a chromosome, including the centromere, telomeres and origins of replication Describe the functional organisation of chromosomes territories in the interphase nucleus and the experimental approaches used to study them Understand and be able to define what is meant by the functional compartmentalisation of the nucleus Describe the structure and function of the nucleolus, as well as other proposed nuclear compartments and the experimental approaches used to study them Describe the nuclear envelope and the functions of the nuclear pore
-
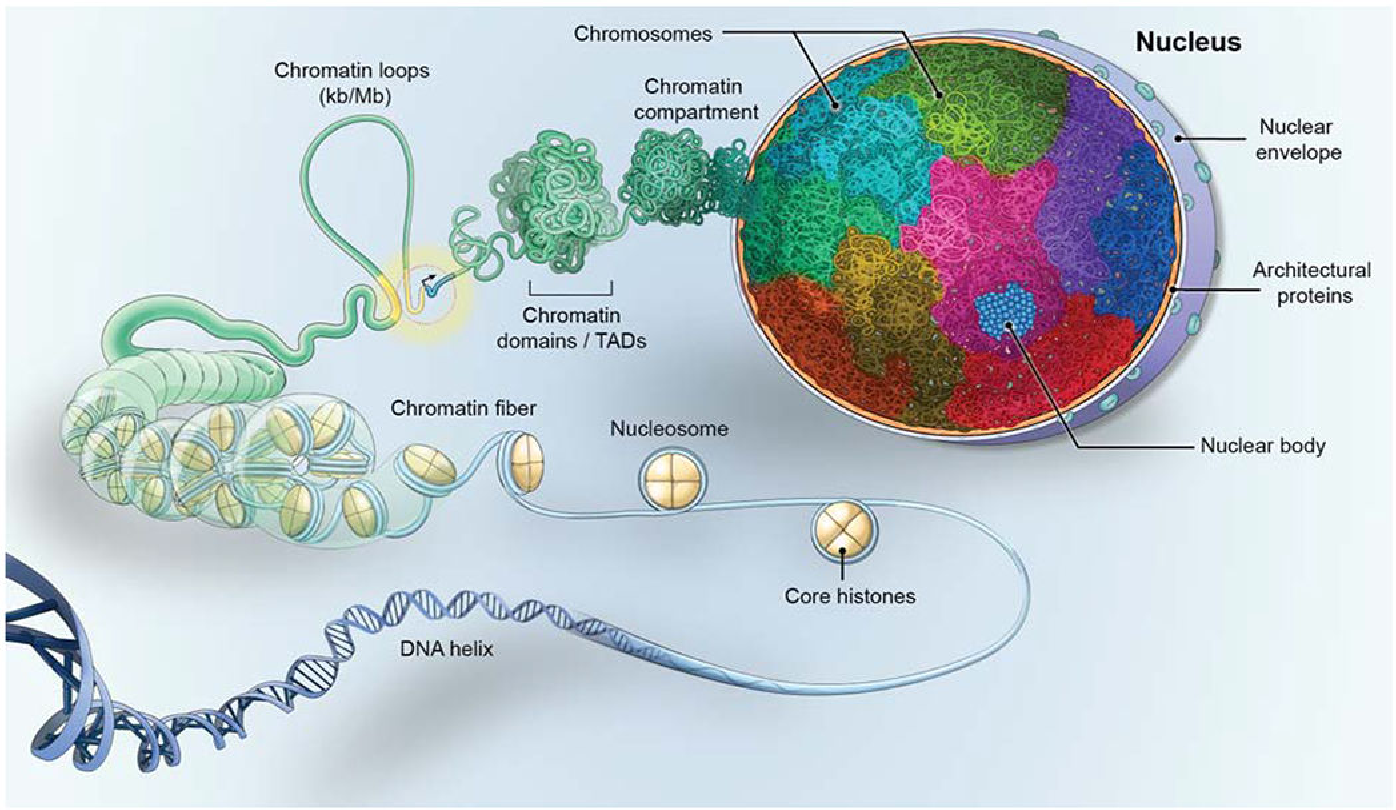
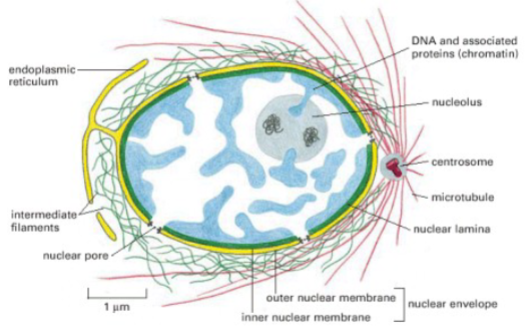
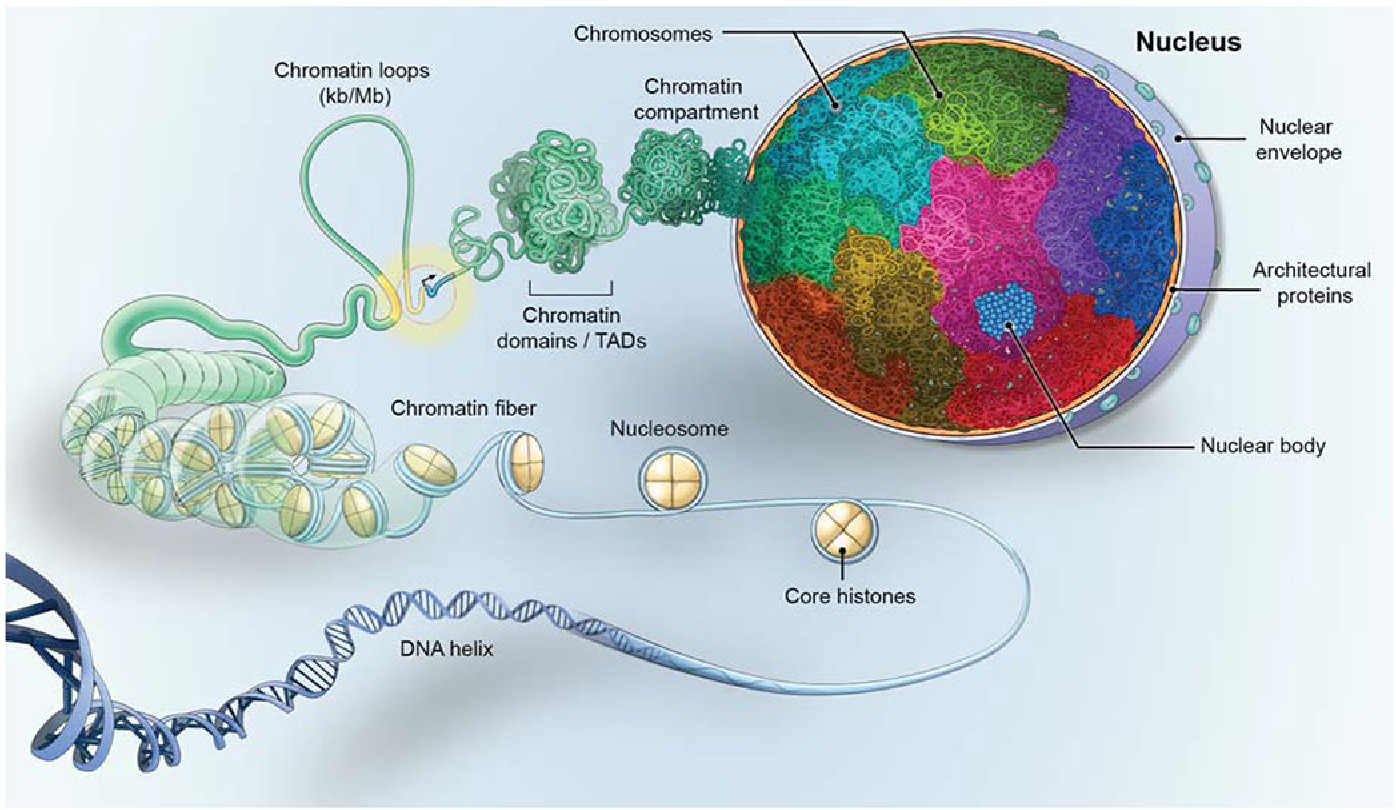
Picture demonstrating the structure of the nucleus
-
Subnuclear compartments exist despite the absence of what?
Internal membranes
-
What do Centromeres do? (2)
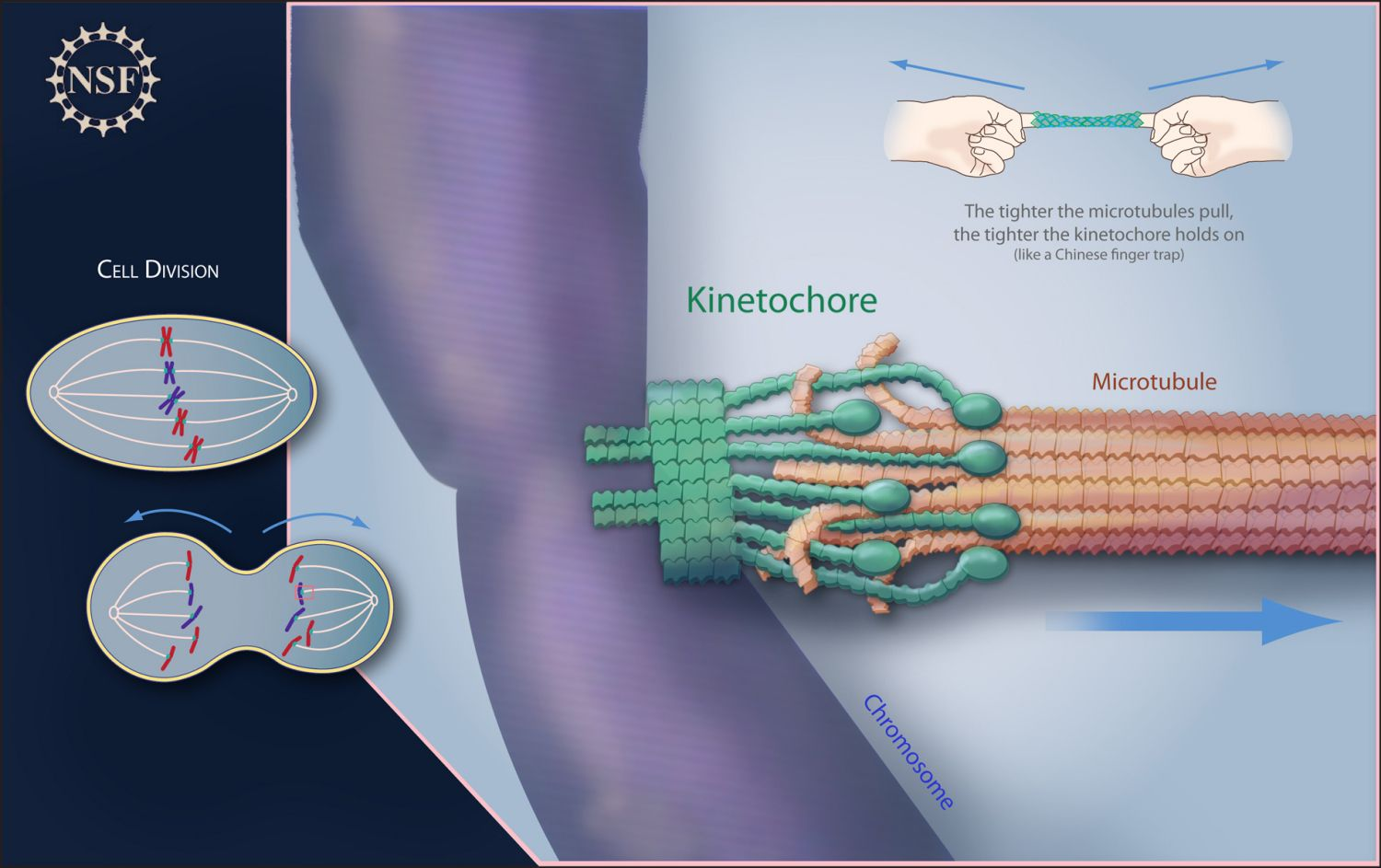
-Locks sister chromatids together
-Attachment site for chromosomes to the mitotic spindle via a protein structure called the kinetochore
-
What are Centromeres? (2) (in context of DNA)
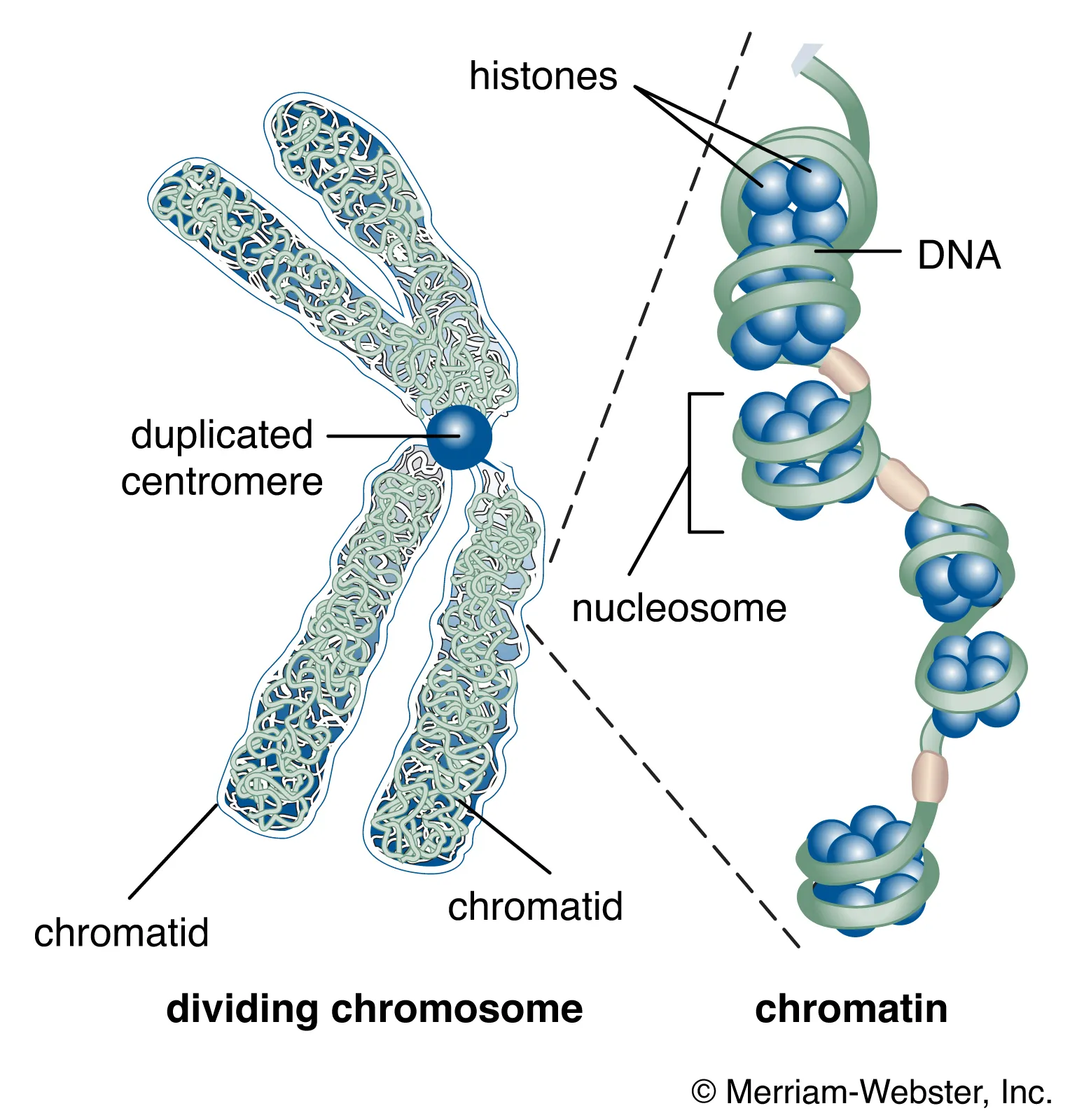
• Megabases of repetitive DNA, major component is the alpha satellite DNA in humans
• All chromosomes have different satellite DNA configurations
-
State some information about Telomeres/end of chromosomes:
• The end of the chromosomes is made up of a tandem repeat (TTAGGG in human)
• With each round of replication, we lose a little bit of the end of our chromosome – the end replication problem
• These repeats are lost as the cell divides
-
What is the Hayflick limit?
Is the number of times, a cell can divide before it dies
-
Explain the telomere end replication problem
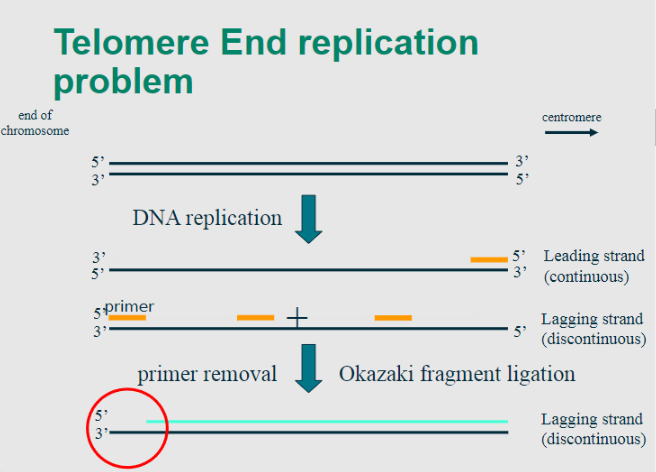
-In the process of DNA replication, the enzyme DNA polymerase synthesizes new DNA strands in the 5' to 3' direction.
-However, due to the nature of linear DNA molecules, the synthesis of the lagging strand occurs discontinuously, creating short fragments known as Okazaki fragments.
-This poses a challenge for replicating the very ends of linear chromosomes.
-
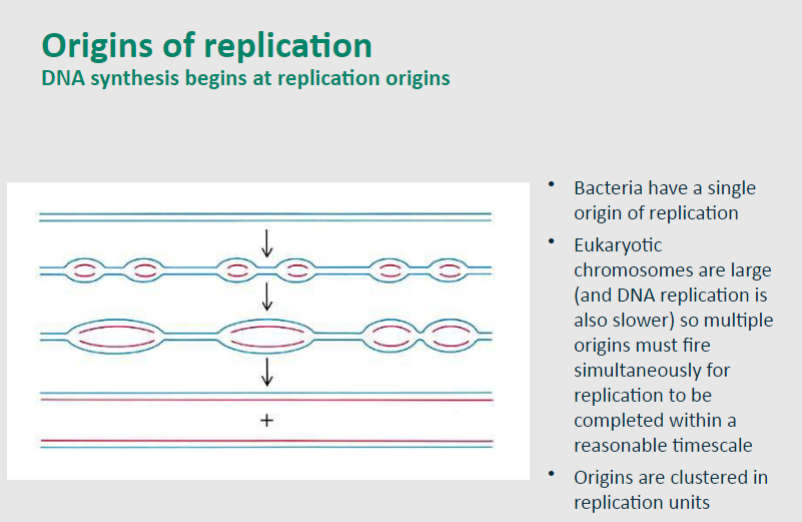
Picture demonstrating how replication origins form:
-
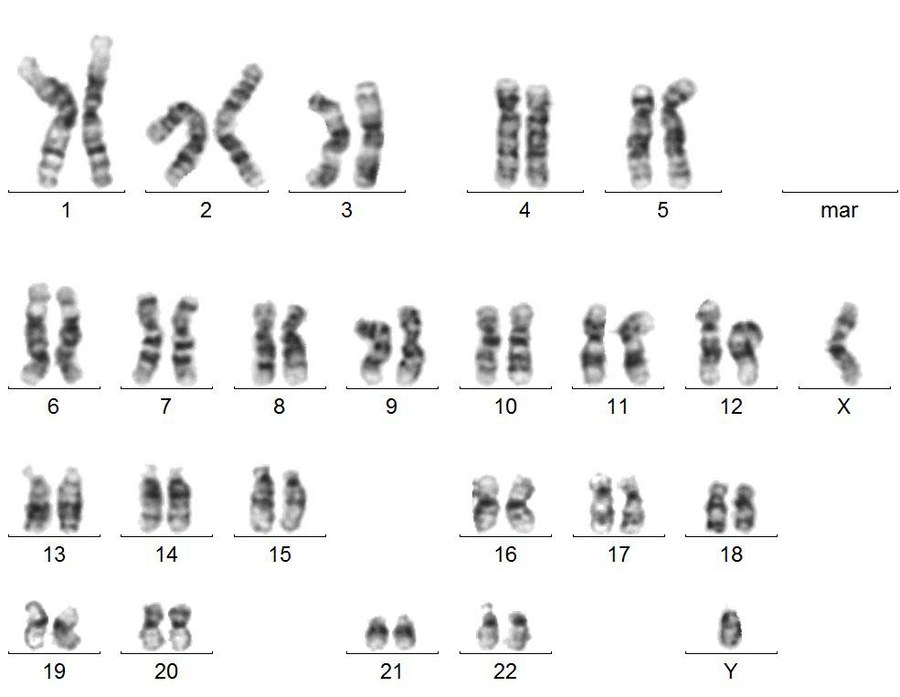
What is Karyotyping?
•Karyotyping is a laboratory technique used to visualize and analyse the number and structure of chromosomes in a cell.
•This process involves arranging and photographing an individual's chromosomes, typically obtained from cells in the metaphase stage of cell division, when chromosomes are most condensed and visible.
-
What is G Banding?
•G-banding is a chromosome banding technique that involves partially digesting chromosomes and staining them with the chemical stain Giemsa.
•The resulting banding pattern reveals alternating dark and light bands along the chromosomes.
•The interpretation of G-banding patterns provides information about the distribution of gene-rich and gene-poor regions on chromosomes.
-
What are G Dark bands?
•G-dark bands appear as dark-staining regions on the chromosomes.
•These bands are often gene-poor and tend to be rich in heterochromatin.
•Heterochromatin is a condensed, tightly packed form of chromatin that is transcriptionally inactive and typically contains repetitive DNA sequences.
-
What are G Light bands?
•G-light bands appear as light-staining regions on the chromosomes.
•These bands are often gene-rich and tend to be rich in euchromatin.
•Euchromatin is a less condensed, more loosely packed form of chromatin that is transcriptionally active
-
What is the Spatial Location of Gene-rich vs Gene-poor Chromosomes?
-Gene-poor at the periphery
-Gene-rich in the interior of the nucleus.
-
Genes can dynamically loop out in response to what activation?
Transcriptional activation
-
Nuclear compartments consist of subnuclear compartments without....?
Internal membranes
-
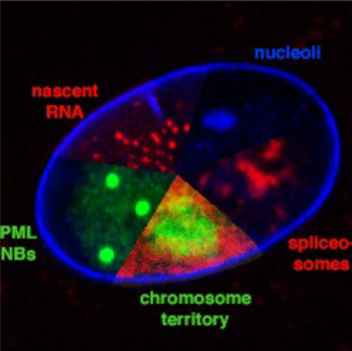
Examples of these subnuclear compartments are? (name 5)
Examples:
-chromosome territories
-replication factories
-transcription factories
-nucleoli
-PML nuclear bodies
-
State some information about the nucleolus:
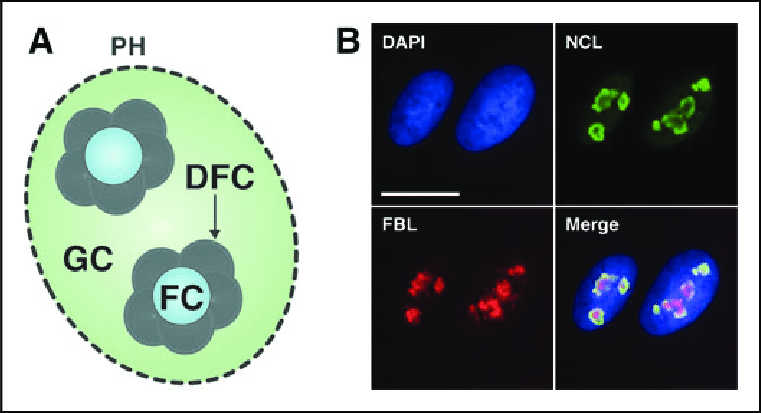
•Largest substructure for ribosomal subunit production.
•Three distinct zones: FC (Fibrillar Center-ribosomal RNA genes), DFC (Dense Fibrillar Component-rRNA transcripts), GC (Granular Component-processing and assembly).
•Self-organizing around Nucleolar Organizing Regions (NORs) with rRNA genes.
-
What is the Fibrillar Center (FC)?
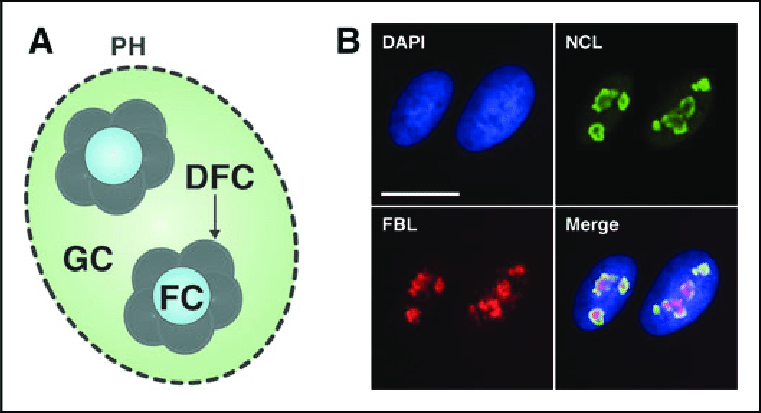
•Location of Ribosomal RNA (rRNA) Genes:
•These genes are responsible for producing the precursor rRNA molecules.
-
What is the Dense Fibrillar Component (DFC)?
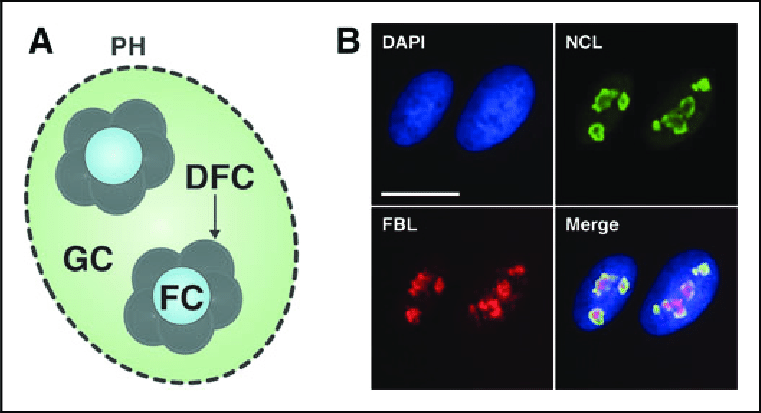
•Site of rRNA Transcription:
•This involves synthesizing larger rRNA molecules that need further processing.
-
What is the Granular Component (GC)?
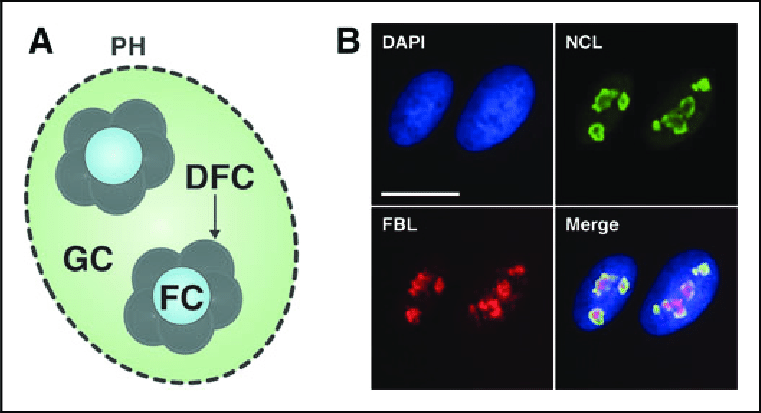
Processing and Assembly:
•Where the precursor rRNA molecules are cleaved and associated with ribosomal proteins to form the 40S and 60S ribosomal subunits
-
What are Nucleolar organising Regions (NORs)?
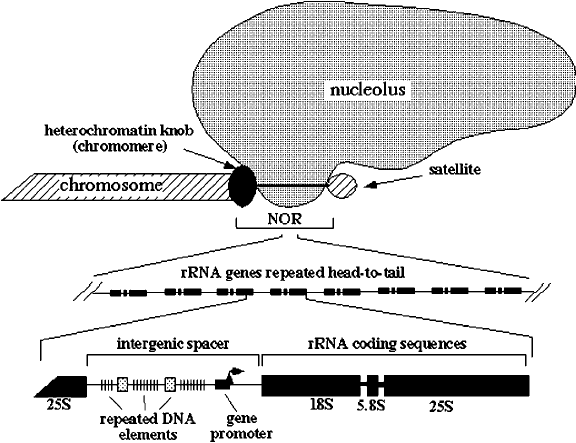
•Nucleolar Organizing Regions (NORs): These are specific chromosomal regions that contain clusters of rRNA genes. •Typically, NORs are located on acrocentric chromosomes.
-
What is the Self-Organizing feature?
•Self-Organizing Feature: The nucleolus forms around these NORs. As transcription of rRNA genes takes place
•The nucleolus self-organizes, creating the distinct zones (FC, DFC, GC) dedicated to different steps in ribosomal subunit production.
-
What is the composition of Splicing Speckles? (2, and state what these parts do)
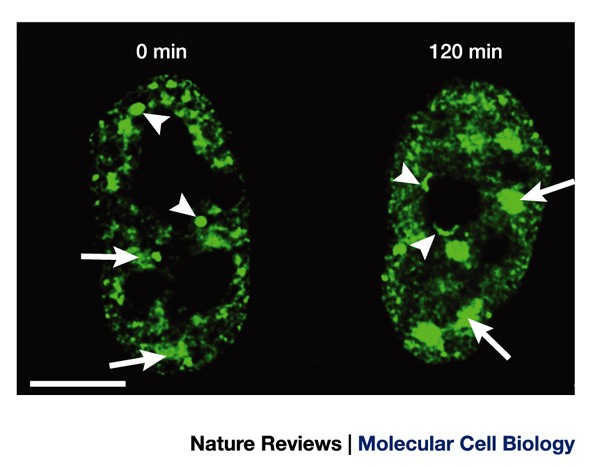
Splicing Factors:
•Splicing speckles are enriched with proteins known as splicing factors.
•They remove introns from precursor mRNA molecules to produce mature mRNA.
mRNA Processing Factors:
•Splicing speckles also contain various proteins involved in mRNA processing.
•They participate in other stages of mRNA maturation, such as capping, polyadenylation, and nuclear export.
-
What is the function of Splicing Speckles? (2 reasons, elaborate on each)
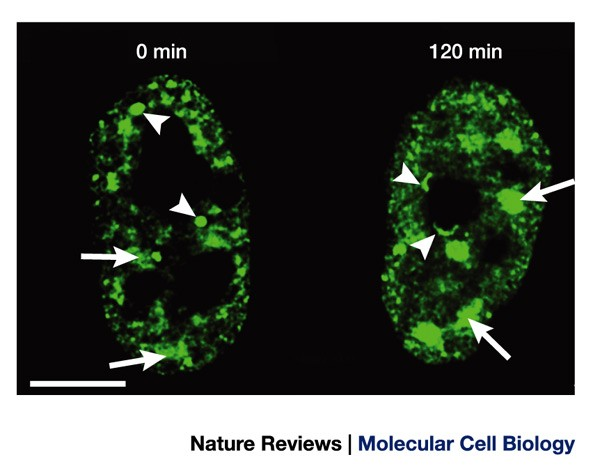
Storage and Assembly:
•Splicing speckles are believed to function as storage and assembly compartments for splicing factors.
•These factors may accumulate in speckles when not actively engaged in splicing processes.
•The speckles serve as reservoirs where splicing factors can be stored before being transported to sites of active transcription when needed.
Regulation of Splicing Factor Access:
•Changes in cellular conditions, such as inhibition of transcription (and thereby splicing) or an increase in splicing activity,
•Can lead to alterations in the size and composition of splicing speckles.
-
How many splicing speckles are in a cell? And how many proteins localise/associate with them?
20-50 per cell
146 known proteins localise/associate
-
What is replication foci?
In mammalian cells, DNA replication takes place at discrete nuclear sites called replication foci where newly synthesized DNA accumulates
-
State some information (2 points) about replication foci
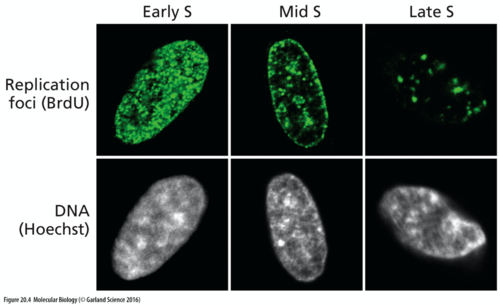
• Different patterns of replication foci have been observed in S phase by many researchers
• No high quality 3D analysis due to limited resolution of optical microscopy
-
State some information about the Replication Factories Hypothesis
• Multiple replicons (DNA replicated from a single origin) must operate in parallel to ensure all genomic DNA is replicated during S phase
• The number of foci is smaller than the number of replicons, leading to the concept of a replication factory
• Factories contain all the enzymes and other factors required to produce two new DNA strands
-
What is satellite DNA?
Highly repetitive DNA consisting of short sequences repeated several times

