Protein Structure and Function: The 3D Structure of Proteins (The Living Cell)
Outcomes Describe the structural and general nature of amino acids and how this influences the chemical properties of a protein Describe primary, secondary, tertiary and quaternary structures of proteins Describe the complexity of proteins and the requirement for some proteins to associate with with other complex molecules, cofactors or ions Define how the structural organisation of the polypeptide chain relates to its functionality Describe using examples the broad categories within which amino acids are grouped with regards their properties Define the bonds and forces that act in the formation and maintenance of structure and their role in the hierarchy of folding Discuss the role freedom of movement of the polypeptide backbone plays in folding and both the rotationary freedom and the limits upon this caused by different factors List the different mechanisms by which protein mis-folding leads to disease Describe using examples different types of protein misfold diseases and the consequences for the fate of different mis-folded proteins
-
Proteins are the fundamental functional molecules of the cell; what are the 5 general functions of proteins?
• Carrier functions
• Metabolic functions
• Form parts of the Cellular machinery
• Make up structural scaffold
• Organelle and Cell communication
-
Are protein polymers?
Yes
+ The basic building blocks of proteins are amino acids+ Amino acids have a generalised structure
-
Describe the structure of Amino Acids: (4)
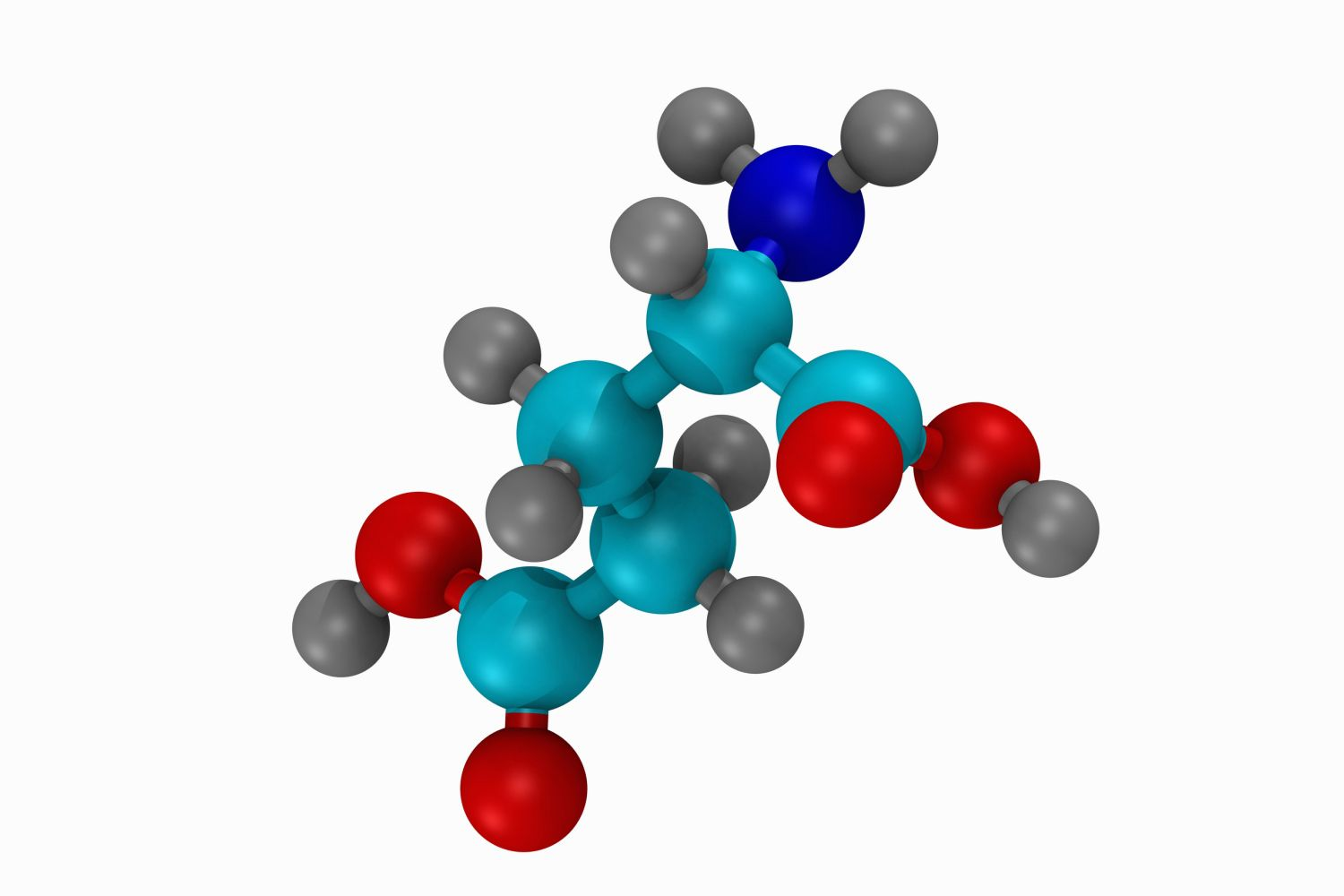
• Tetrahedral arrangement of atoms with the alpha carbon at its centre
• Carboxyl and amine groups attached to a central carbon
• A variable side chain represent by R confers the specific physicochemical properties
• And a Hydrogen
-
What are the ionic properties of AA (Amino Acids)?
• Amine and Carboxyl groups readily ionise
• Acidic and basic groups - AA are in equilibrium with the surrounding environment
-
What does 'ionic' even mean?
-A type of chemical bond formed by the electrostatic attraction of oppositely charged ions.
-
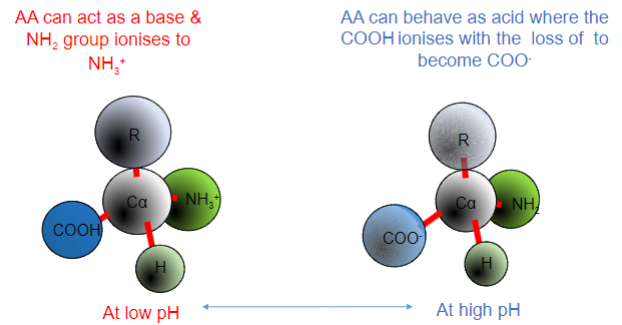
How does the AA (groups) react at low and high pHs?
-
The arrangement of atoms gives rise to two Isomers... what are the names?
D and L
-
Elaborate on the D isomers of Amino Acids:
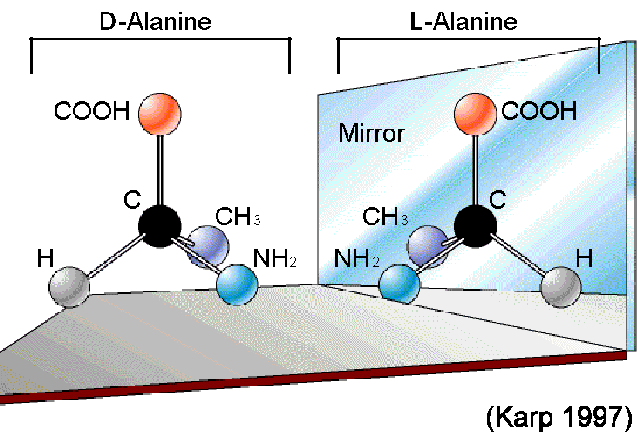
2. D-isomer (dextro or right):
The amino group (NH2) is on the right side in a Fischer projection.
D-amino acids are less common in nature and are not typically found in proteins, although some exceptions exist.
-
Elaborate on the L isomers of Amino Acids:
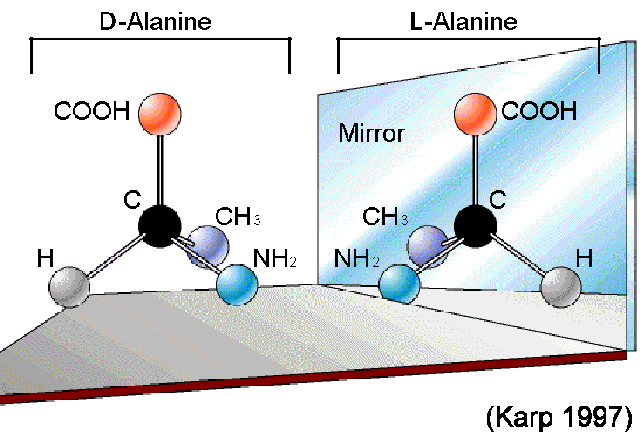
1. L-isomer (levo or left):
The amino group (NH2) is on the left side in a Fischer projection.
Most naturally occurring amino acids in proteins are in the L-configuration
-
How do you describe the arrangement around the alpha carbon atom? Is there an exception?
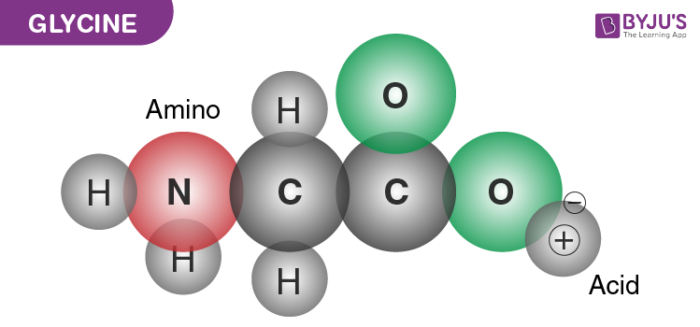
-Asymmetric
-Chiral (exception is Glycine, where R is a hydrogen)
-
Picture outlining how these differences can be seen:
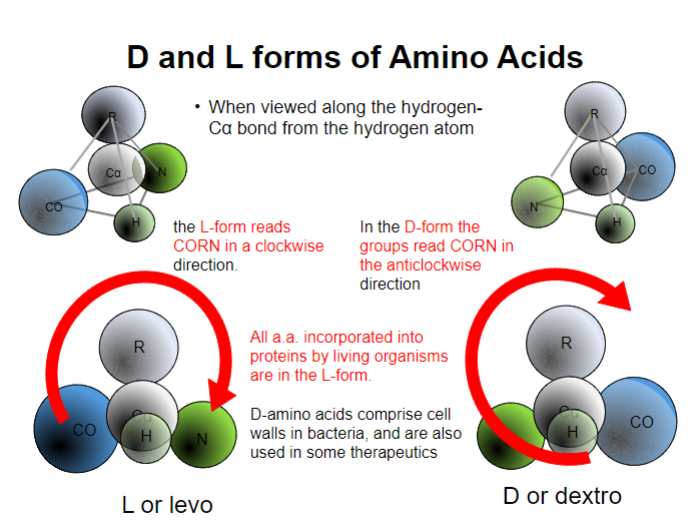
-
AA combine via a what bond? What type of reaction? Releasing what?
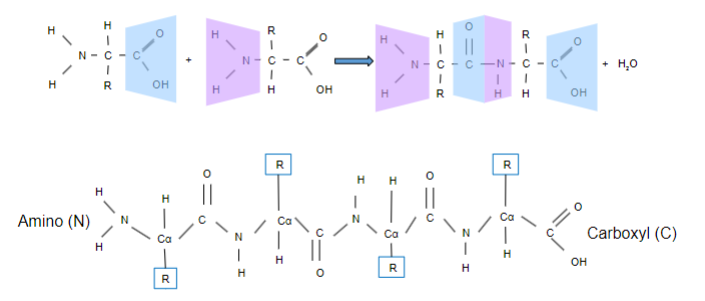
-Peptide
-Condensation
-Releasing H2O
-
What are peptide residues?
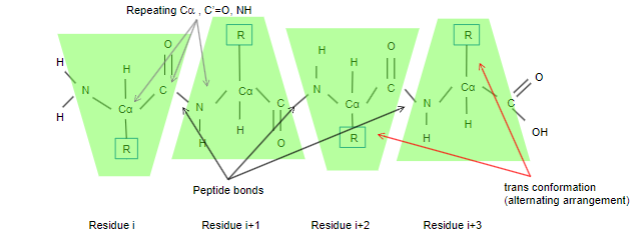
•Each repeating unit of the polypeptide chain is termed a residue
•Each residue consists of an invariable unit comprising
• Alpha carbon, C’=O and NH, group
•The variable side chain R is usually arranged in a trans conformation
• 0.1% peptide bonds have a less energetically favourable cis arrangement
-
What is the Primary structure of Proteins?
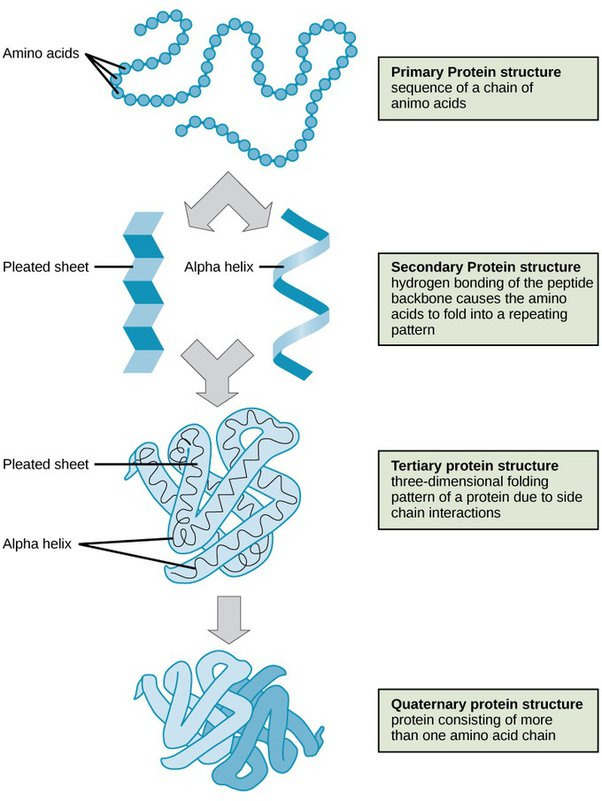
• Covalent bonds forming polypeptide chain – i.e. order of aminoacid residues in the polymer
-
What is the Secondary structure of Proteins?
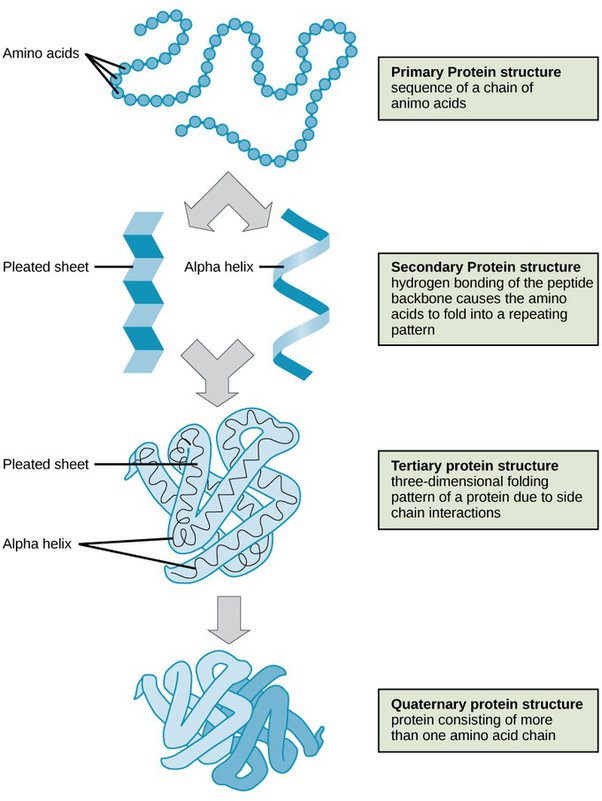
• Regular folded form stabilised by hydrogen bonding, e.g. alpha helices, beta sheets and beta turns
-
What is the Tertiary structure of Proteins?
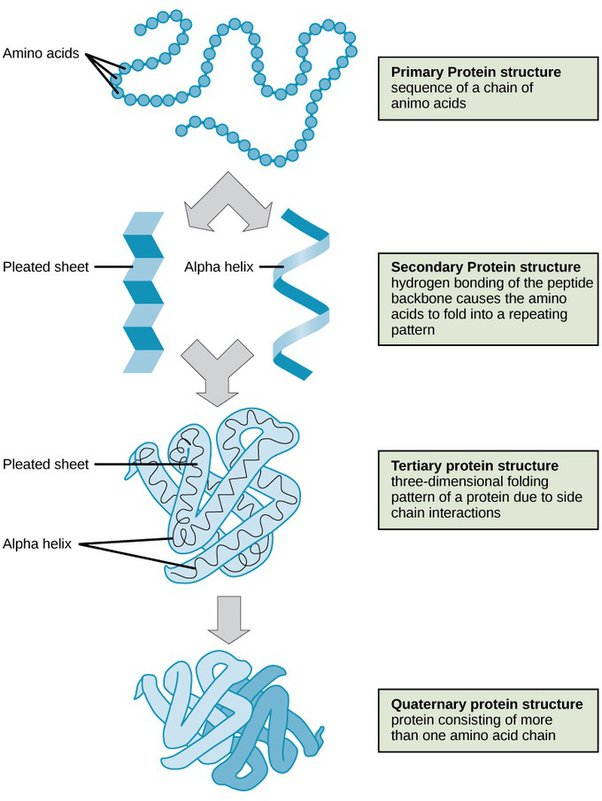
• Overall 3D structure, stabilised by non-covalent bonds and forces, and sometimes by intra-chain covalent bonds
-
What is the Quaternary structure of Proteins?
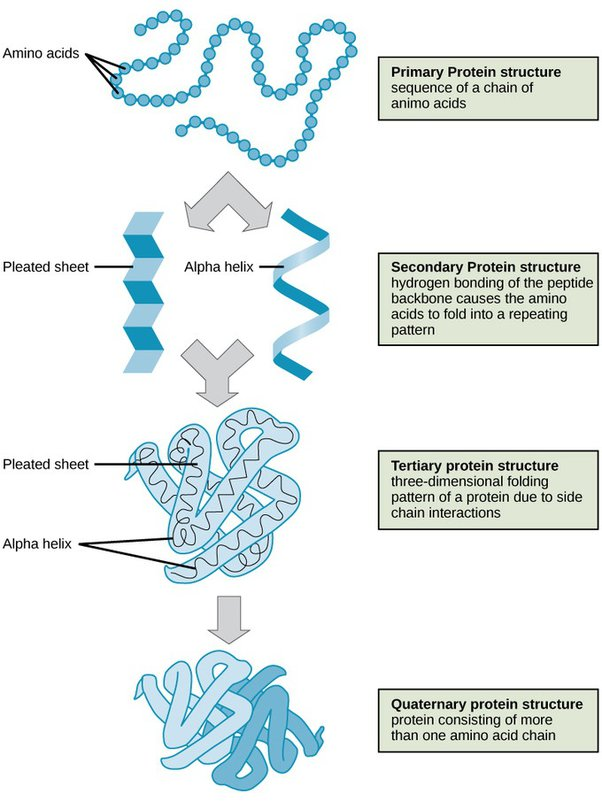
• Organisation of polypeptides into assemblies, stabilised by non-covalent bonds and forces (as for Tertiary) and sometimes interchain covalent bonds
-
Secondary Structure includes beta sheets and strands, what are they?
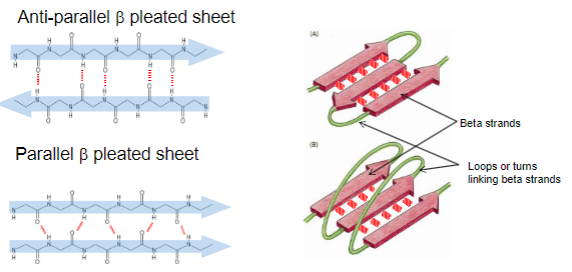
•beta sheets (are formed from hydrogen bonding) between adjacent beta strands, either in antiparallel or parallel arrangement
•beta strands are connected by loops or short turns, depending upon their arrangement
-
Secondary Structure includes alpha helixes, what are they?
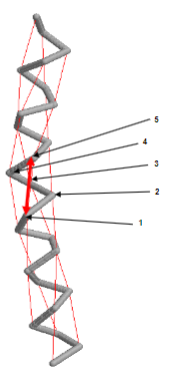
• Right handed helix
• stabilized by H-bonds
• H-bonds are 4 residues apart between residue 1 and 5
• 3.6 residues per turn or 0.54 nm per turn
-
Most functional proteins are formed from two or morefolded......?
-folded polypeptides that combine to form the mature protein.
-
Some Proteins require cofactors for their function, others for their folding: Can you list some examples? (3)
• Heme of Haemoglobin,
• Nicotinamide adenine diphosphate (NAD)
• Flavin adenine dinucleotide (FAD)
- Organic cofactors are sometimes called coenzymes, but other proteins might include ions- e.g. Zn2+ Mg2+ Ca2
-
Water-soluble proteins usually have what shape?
Globular
-
Name some characteristics of water-soluble proteins? And some examples of water-soluble proteins
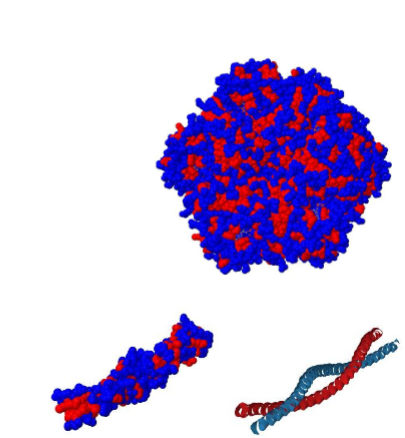
-Hydrophobic Core (Residues): Globular proteins have a compact structure with a hydrophobic core, where nonpolar amino acid side chains cluster together to minimize contact with water.
-Hydrophilic Surface (Residues): The exterior surface of globular proteins is hydrophilic, allowing them to interact favourably with the surrounding aqueous environment
Examples:
Some Water soluble proteins:
• May assemble into filaments (e.g. actin) or tubes (e.g. tubulin) or coiled-coils (cortexillin)
-
This organization of residues is reversed in membrane proteins: Elaborate
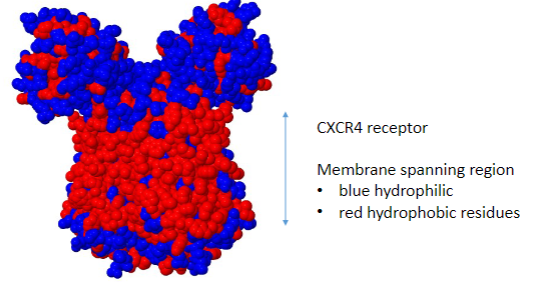
• Membrane spanning regions have externally located hydrophobic residues that interact with the membrane lipids.
• They may have hydrophilic central channels.
-
Properties of Amino Acids are determined by their structure, How?
1)If they are CHARGED!
• May be subdivided into acidic or basic (alternatively positive or negative)
+Acidic = -ve charge
+Basic = +ve charge
2)Charged amino acids are also polar in nature
3)Sometimes the sulphur containing amino acids are a separate group
4)Glycine is sometimes considered Nonpolar, sometimes it’s a separate group
5)Histidine is sometimes also considered as a charged basic amino acid
-
What are the properties associated with each of the groups?
– Carboxyl groups COO- ⇾ charged or acidic
– Amine groups NH3+ → charged or basic
– 2o (secondary) Amine NH and Carbonyl C=O groups →polar
– Hydroxyl OH → polar
– Hydrocarbon → Non-polar Hydrophobic
-
Where is the rotational freedom of the polypeptide found?
• The rotational freedom within a polypeptide is found around alpha carbon, NOT the peptide bond
-
Polypeptides adopt a structure based on Energy minimisation: Elaborate on ∆G.......What is the formula?
The equation for ΔG is:
ΔG=ΔH−TΔS
where:
ΔH is the change in enthalpy (heat absorbed or released during the reaction),
T is the temperature in Kelvin,
ΔS is the change in entropy.
The sign of ΔG determines the spontaneity of a reaction:
If ΔG is negative, the reaction is spontaneous (exergonic), releasing energy.
If ΔG is positive, the reaction is non-spontaneous (endergonic), requiring an input of energy.
If ΔG is zero, the system is at equilibrium.
-
Non-covalent bonds have what strength compared to covalent bonds? However, what?
-Non-covalent bonds have 1/20th strength of covalent bonds
-However...the overall contribution of non-covalent bonds are significant because non-covalent bonds are far larger
-
What are the four Non-covalent bonds and forces? Elaborate on each force
1. Charge or electrostatic attractions
• falls off exponentially as distance increases, affected by electrostatic environment (aqueous environment)
2. Hydrogen bonds
•(transient bonds, similar in some respects to covalent bonds)
3. Van der Waals attractions – dipole These weak forces occur between two atoms• determined by their fluctuating charge
• Van der Waals forces are induced by proximity of molecules
4. hydrophobic interactions
• (water is a polar molecule) hydrophobic interactions minimise disruption of water network – i.e. the fourth weak force Non-covalent bonds and forces
-
Covalent bonds Disulphide bonds
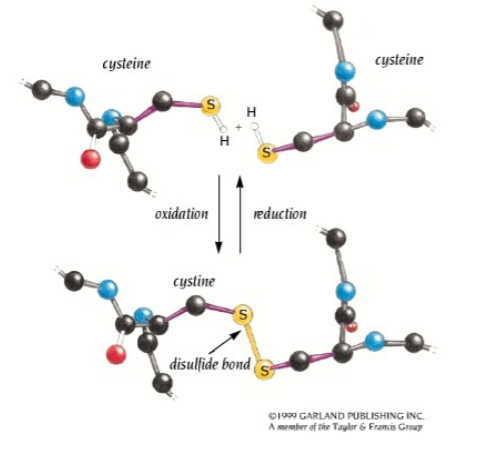
-Disulphide bonds form between the side chains of two cysteine residues
• Bonds form in an oxidative reaction
• The SH groups from each cysteine cross-link.
• Usually occurs in distant parts of the primary sequence but adjacent in the three-dimensional structure.
• Can form on the same (intra-chain) or different (interchain) polypeptide chains, e.g. insulin left
-
When proteins are misfolded, disease occurs: How (generally)? Name some diseases caused by protein misfolding
• The function of the misfolded protein is almost always lost or reduced
• Misfolded proteins tend to self-associate and form aggregates
• Huntingtin Htt (Huntington’s)
• Amyloid-beta Ab (Alzheimer’s)
• prion protein (PrPSc) Creutzfeldt–Jakob disease and variant CJD
• alpha-synuclein (Parkinson’s disease)
• Serum amyloid A (AA amyloidosis)
• islet amyloid polypeptide IAPP (Type 2 Diabetes) and many more
• Other misfolded proteins result in cellular processing that lead to their degradation
• Cystic fibrosis
-
When proteins are misfolded, disease occurs: How (specifically)? (6)
1. somatic mutations in the gene sequence, leading to the production of a protein unable to adopt the native folding
2. errors in transcription or translation leading to the production ofmodified proteins unable to properly fold
3. Failure of the folding machinery
4. mistakes of the post-translational modifications or in trafficking ofproteins
5. Structural modification produced by environmental changes
6. induction of protein misfolding by seeding and cross-seeding by other proteins
-
What is the Protein folding and misfolding and disease Amyloid hypothesis?

This hypothesis proposes that the aggregation and accumulation of Aβ (Amyloid beta) peptides, particularly in the form of amyloid fibrils, play a central role in the development and progression of Alzheimer's disease
In Alzheimer's Disease:
•The 40 amino acid β-Amyloid (Aβ) peptide accumulates.
•Misfolding of β-Amyloid (Aβ) results in a flat arrangement forming a beta sheet.
•Polymerization forms fibrils of misfolded protein known as amyloid fibrils.
•Stacked beta sheets of β-Amyloid (Aβ) are stabilized by side chains that interdigitate (weave together)
-
What is the Protein folding and misfolding Cystic fibrosis hypothesis?
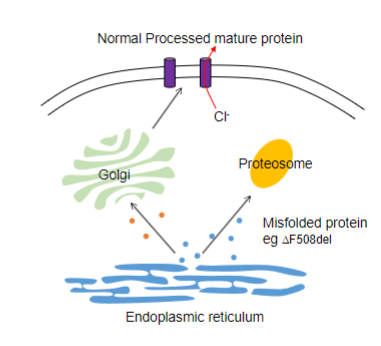
Common Mutation:
In Cystic fibrosis, the prevalent mutation is the deletion of Phenylalanine at residue 508 of the CFTR (Transmembrane conductance regulator) gene.
DF508del Mutation and Misfolding:
The DF508del mutation leads to the misfolding of the CFTR protein, occurring while the protein is still within the ER.
Recognition by Cellular Machinery:
The misfolded CFTR protein is recognized by the cellular machinery responsible for identifying and processing misfolded proteins.
Ubiquitination and Proteasomal Degradation:
-This results in ubiquitination, trafficking to the proteasome and degradation
-
What is Seeding?
Seeding: Process where misfolded proteins act as templates, inducing the misfolding of normal proteins.
-
What is Cross-Seeding?
Cross-Seeding: Involves misfolded proteins from one type encouraging the misfolding of a different protein type, potentially amplifying the protein misfolding cascade.

