Linkage Analysis (Genomics) UNFINISHED
Linkage Analysis This lecture is designed to teach you the basic principles of linkage analysis and how it is used to identify Mendelian disease genes. To start, we will review the mechanisms of genetic variation and why it is important. Next, we will look at the genetic concepts that are important for linkage analysis, i.e. homologous recombination and independent assortment. Finally, you will be shown different genotyping methods used for linkage analysis and how statistical analysis can be applied to generate a LOD (logarithm of the odds) score. Learning Outcomes On successful completion of the lecture, students should be able to: Explain homologous recombination and genetic linkage Describe how genetic disease can be classified by the number of genes required to cause the disease Describe the difference between genetic maps and physical maps of the genome Describe the principles of linkage analysis and why it is useful Describe the differences between the two types of genetic marker routinely used in linkage studies Briefly describe the genotyping technologies used in linkage analysis Describe what a haplotype is and how it is built Briefly describe the statistical approaches used in linkage analysis, with specific emphasis on the logarithm of the odds (LOD) score Summarise how linkage analysis can be combined with whole exome sequencing to identify a disease-causing variant
-
What is genetic variation, and what are its causes? (3)
Genetic variation refers to differences in the DNA sequence between individuals in a population.
Variation can be inherited or arise from environmental factors, such as drug exposure or radiation.
Genetic variation can affect the amino acid sequence of proteins, gene regulation, physical appearance (e.g., eye color, disease risk), or have silent/no apparent effects.
-
What are the mechanisms of genetic variation? (2)
Mutation refers to changes in the DNA sequence that can lead to variations, while polymorphism refers to variations that are common in the population.
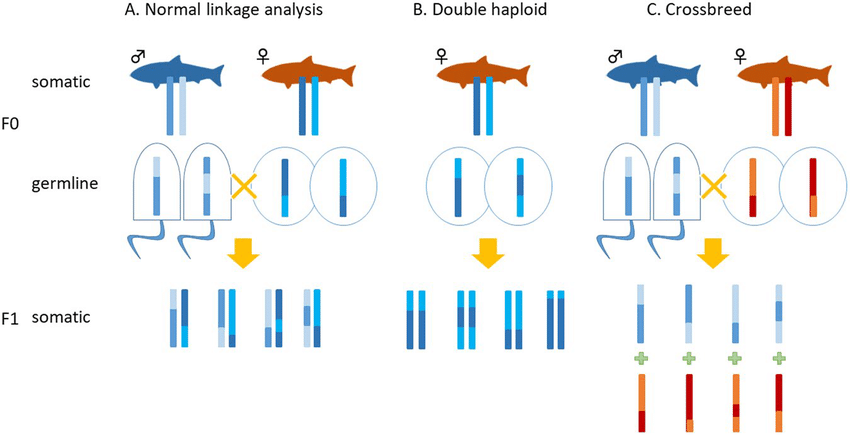
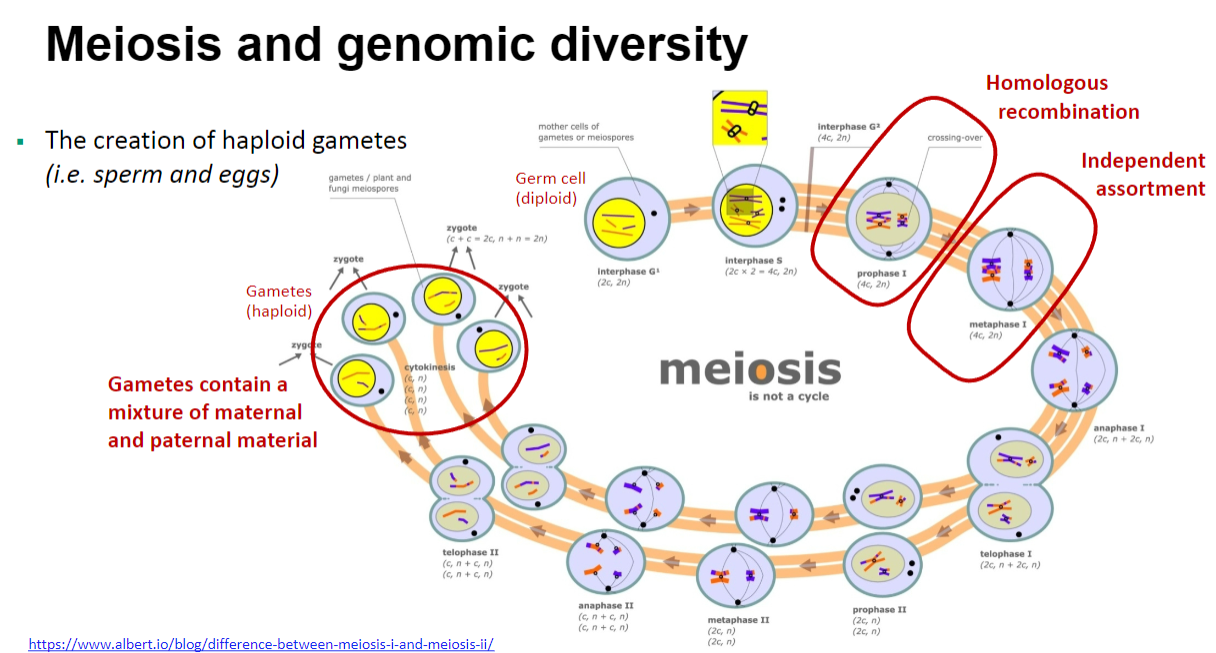
Homologous recombination and independent assortment contribute to genetic variation during meiosis, leading to diverse genetic combinations.
-
What is the difference between Mendelian (monogenic) and Complex (polygenic) diseases? (2)
Mendelian diseases are caused by mutations in a single gene (monogenic), while complex diseases are influenced by multiple genes and environmental factors (polygenic).
Linkage analysis is typically used for Mendelian diseases, whereas association analysis is often used for complex diseases.
-
What are genetic maps and physical maps? (2)
Genetic maps are based on the frequency of recombination between markers and provide a relative position of genes.
Physical maps represent the actual physical distance between genes on a chromosome, measured in base pairs.
-
What are haplotypes, and how are they used in linkage analysis? (2)
Haplotypes are combinations of alleles at multiple loci that are inherited together from a single parent.
They are used in pedigree analysis to track the inheritance of genetic traits and help identify genetic markers associated with diseases.
-
What statistical approaches are used in linkage analysis? (1)
LOD (Logarithm of the Odds) scores are generated through statistical approaches to assess the likelihood of linkage between genetic markers and traits in pedigree analysis.
-
What is homologous recombination and its significance? (3)
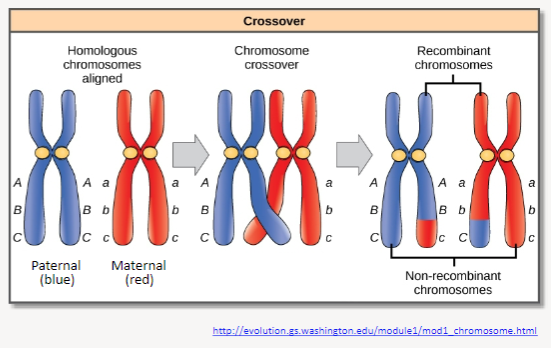
Definition: Homologous recombination involves crossing over, which is the reciprocal breaking and re-joining of homologous chromosomes during meiosis.
Result: This process results in the exchange of chromosome segments between homologous chromosomes.
Significance: It leads to new allele combinations, increasing genetic diversity within a population and contributing to evolution.
-
Picture demonstrating Meiosis and genomic diversity:
-
What are key genetic terms and their definitions? (6)
Genotype: The genetic makeup of an individual.
Phenotype: The physical expression of the genetic makeup.
Alleles: Alternative versions of a gene found at the same locus on homologous chromosomes.
Inheritance: An organism inherits two alleles for each characteristic, one from each parent; these alleles can be the same (homozygous) or different (heterozygous).
Homozygous Genotype: A genotype with identical alleles for a specific locus.
Heterozygous Genotype: A genotype with two different alleles for a specific locus.
Haplotype: A group of alleles that are inherited together from a single parent (the prefix ‘haplo’ means single).
-
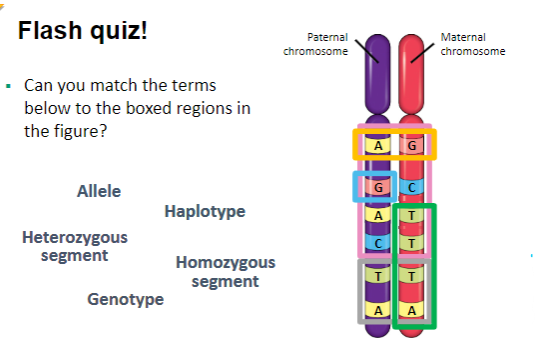
Can you match the terms
below to the boxed regions in
the figure?

-
What is the difference between homozygosity and heterozygosity? (3)
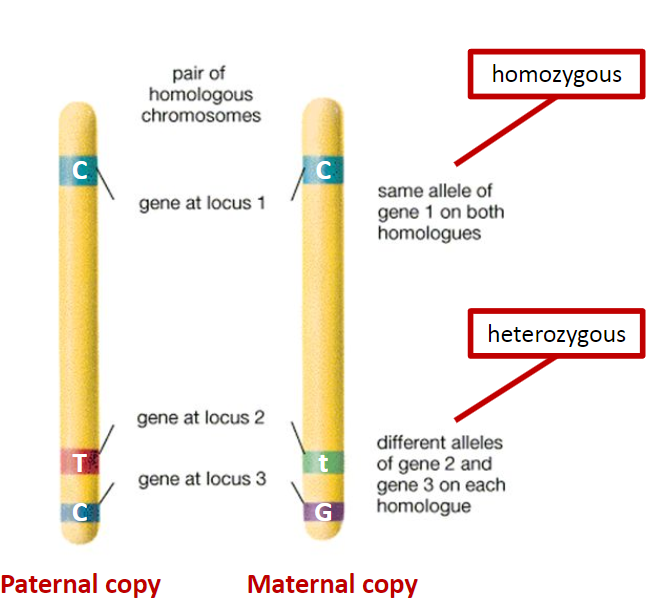
Chromosome Pair: Homozygosity and heterozygosity refer to the alleles at the same loci on homologous chromosomes.
Homozygosity: If the alleles at a given locus are the same, the individual is homozygous for that trait.
Heterozygosity: If the alleles at a given locus are different, the individual is heterozygous for that trait.
-
What are the classifications of genetic diseases? (3)
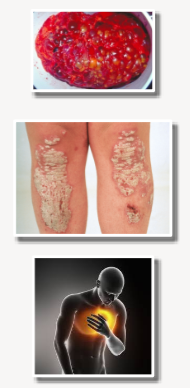
Mendelian/Monogenic: Diseases caused by a single gene with little or no environmental influence (e.g., Polycystic Kidney Disease (PKD)).
Non-Mendelian/Polygenic/Complex: Diseases or traits resulting from the cumulative effect of many different genes, each contributing a small individual impact (e.g., psoriasis).
Multifactorial: Diseases or traits that arise from the interaction of multiple genes and often involve multiple environmental factors (e.g., heart disease).
-
What are the characteristics of rare genetic diseases in relation to their genetic classification? (4)

Monogenic Diseases: Rare genetic diseases are typically monogenic, meaning they are caused by a single gene.
Single Gene Impact: In monogenic diseases, one gene corresponds to one disease, often due to a single rare variant with a significant effect on protein function.
Polygenic Diseases: Complex diseases, on the other hand, are polygenic, meaning they involve many genes contributing to one disease.
Common Variants: These complex diseases are caused by the cumulative impact of common variants, each exerting a small effect on the overall condition.
-
Picture demonstrating Genomic variation and gene mapping in disease:
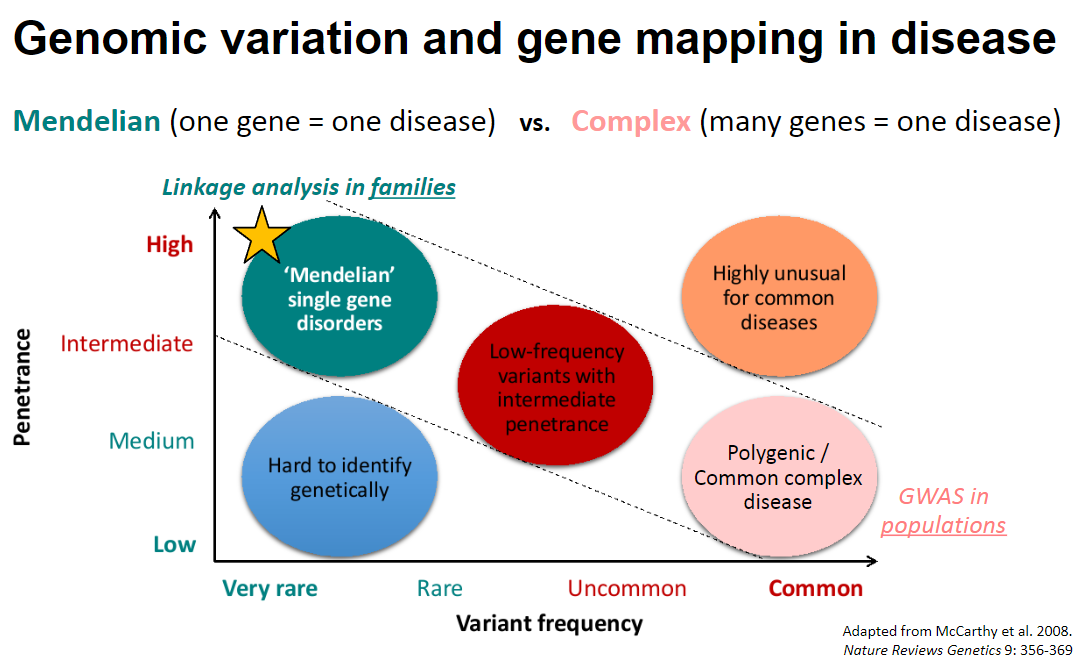
-
What is linkage analysis? (3)
Definition: Linkage analysis is a method used to map the location of a disease gene within the genome.
Linkage Concept: The term 'linkage' refers to the assumption that two genetic elements are physically linked to each other.
Purpose: This technique helps in identifying the chromosomal location of genes associated with specific diseases, aiding in genetic research and diagnosis.
-
What are the assumptions of linkage analysis? (3)
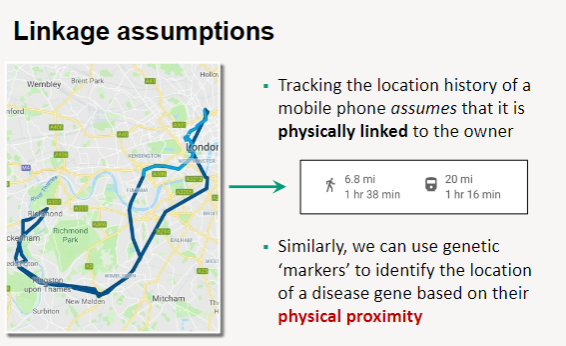
Physical Linkage: Linkage analysis assumes that genetic markers are physically linked to each other, similar to how a mobile phone is linked to its owner.
Tracking Location: Just as the location history of a mobile phone can be tracked based on its connection to the owner, genetic markers can track the location of a disease gene.
Proximity Utilization: By using genetic markers that are in close physical proximity to a disease gene, researchers can identify and map the gene's location within the genome.
-
What is the importance of maps in navigation? (3)
Contextual Orientation: Maps provide context, helping individuals orient themselves within a space and understand their surroundings.
Distance Calculation: They allow users to calculate the distance between landmarks, facilitating efficient navigation.
Reference Points: For example, tube stations serve as reference points or ‘landmarks’ that aid in navigating through a city.

