Mutation
advertisement

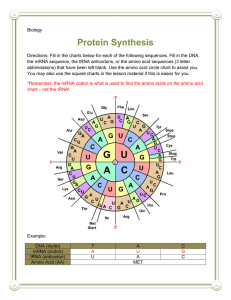
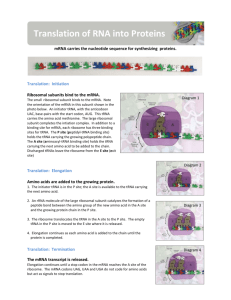
Announcements 1. Homework - problem set 5 - due this Friday 2. Reading Ch. 14: Skim btm 391 -top 397. Skip rest of 397- 403. Review of Last Lecture I. tRNA and the genetic code II. Transcription - prokaryotes III. Transcription - eukaryotes Outline of Lecture 25 I. RNA processing in eukaryotes II. Translation of mRNA into protein - tRNA and ribosomes III. Three steps of translation IV. First evidence that proteins are important to heredity V. One gene- one enzyme hypothesis I. RNA Processing in Eukaryotes STABILITY STABILITY Introns and Exons Eukaryotic vs. Prokaryotic Transcription • In eukaryotes, transcription and translation occur in separate compartments. • In bacteria, mRNA is polycistronic; in eukaryotes, mRNA is usually monocistronic. – Polycistronic: one mRNA codes for more than one polypeptide – moncistronic: one mRNA codes for only one polypeptide • 3 RNA polymerases in euk., 1 in prok. • Binding of Basal Transcription Factors required for euk. RNA Pol II binding. • “Processing” of mRNA in eukaryotes, no processing in prokaryotes II. Structure: Unusual Bases Found in tRNA Function of Unusual Bases • Created post-transcriptionally. • Purpose is sometimes to allow for promiscuous basepairing: Inosine in the 1st “wobble” position of anticodon can bind to 3rd U, C or A in codon. • This means that fewer different tRNAs are required. • Others play a structural role. tRNA Structure Aminoacyl tRNA synthetase Aminoacyl tRNA Synthetases • Enzymes which bond specific amino acids to their cognate tRNAs. • There are 20 different synthetases, one for each amino acid. • Covalent linkage through an ester bond (amino acid activation) requires ATP. • tRNA linked to amino acid is charged. Ribosome Structure S = Svedberg, a measure of sedimentation in centrifuge Ribosome Binding Sites: A, P, E III. Translation has 3 Steps, Each Requiring Different Supporting Proteins • Initiation – Requires Initiation Factors • Elongation – Requires Elongation Factors • Termination – Requires Termination Factor Overview of Prokaryotic Translation Initiation: 1. Binding of initiation factors to small subunit. 2. Binding of first tRNA and mRNA to small subunit. 3. Binding of large subunit. Elongation: 1. Binding of next tRNA using EFs at A site. EPA EPA 2. Peptide Bond formation between 2 amino acids. EPA EPA EPA 3. Translocation of ribosome. Termination: 1. Binding of Release Factor to Stop Codon UGA, UAA, UAG. 2. Disassembly EM of Polyribosomes: >1 Ribosome working on the same mRNA Rabbit Hemoglobin mRNA Midgefly Salivary Gland with Nascent Polypeptide Note: occurs in cytoplasm. IV. Inborn Errors of Metabolism Provided First Evidence that Genes Encode Proteins Alkaptonuria is an inherited disorder first described by Garrod (1902) and Willliam Bateson. – Infants have black urine, darkened ears and nose due to homogentisic acid deposits. – Garrod increased the amino acids phenylalanine and tyrosine in the diet and saw increased deposits in affected individuals only. – He concluded that “unit factors control ferments” (genes control enzymes); results ignored for 30 years. Phenylketonuria (PKU) • Autosomal recessive human metabolic disorder, first described in 1934. • 1/11,000 live births, results in mental retardation due to high [Phe] in body fluids. • Homozygotes cannot convert Phe to Tyr, since enzyme phenylalanine hydroxylase is lost. • Treatment: detection in newborns, low Phe diet; prevents mental retardation • Thousands of disorders have been found that result from genetic factors rather than pathogens. Metabolic Pathways for Phe and Tyr tyrosinase Other Metabolic Disorders in the Pathway • Albinism – Autosomal recessive – Results from loss of tyrosinase enzyme in skin, which converts Tyr to DOPA and DOPA to Melanin pigments – Loss of tyrosinase in brain causes Parkinson’s Disease (loss of DOPA+ neurons). • Tyrosinemia – Results from loss of tyrosine transaminase V. Beadle and Tatum: One Gene One Enzyme (Polypeptide) • From mutations in fungus Neurospora • True in many cases, but there are many exceptions: – Some proteins have multiple subunits, each a polypeptide coded by a different gene. – Some genes code for more than one polypeptide, through differential splicing out of introns; e.g. secreted vs. membrane-bound forms of antibody molecules.