Transcription Translation
advertisement

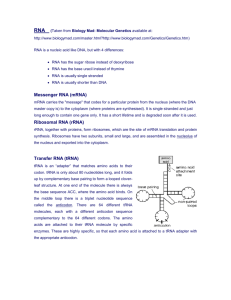
From Gene to Protein PROTEIN SYNTHESIS You Must Know The key terms : Transcription and translation Explain the process of transcription How eukaryotic cells modify RNA after transcription The steps of translation How point mutations change the amino acid sequence of a protein Hint from me as a reader Central Chapter for Molecular Genetics- One of the top 5 to study for AP EXAM Genes Specify Proteins Via Transcription and Translation One Gene one polypeptide hypothesis Each gene codes for a polypeptide which can be or be part of a protein Overview of Transcription/Translation Transcription Translation Synthesis of RNA or mRNA Polypeptide production of under the direction of DNA mRNA is produced Prem-RNA undergoes processing to yield mRNA Prokaryotes- no premRNA polypeptide chain using mRNA transcript that occurs in ribosome Uses triplet code-codon Single strand DNA transcribed to complementary to the original DNA mRNA base triplet codes are called codons, 5’-3’ direction Transcription and translation simultaneously produced Redundant Genetic Code More than 1 codon codes for each of the 20 amino acids Codons are in groups of 3 and are read in a frame Must be read in correct groupings for translation to be successful Prove it!!! Archibald Garrod First to suggest genes dictate phenotype through enzymes Inborn errors of metabolism May be the first to recognize Mendel’s pea characteristics of inheritance apply to humans as well More Proof!!! Beadle and Tatum Bread mold (Neurospora) Bombarded with x-rays to mutate Medium (agar) had 20 amino acids to supplement mutant’s growth even if 1 was missing. Plated these on specific medium to determine what supplements were missing from fungi Nutritional Mutants in fungi Beadle and Tatum Bread mold exposed to xrays Mutants that were unable to survive on minimal media Found 3 classes of arginine deficient mutants Each lacked a different enzyme for catalysis Developed One Gene one Enzyme Hypothesis Central Dogma Universality of the genetic code Nearly universal from simple bacteria to complex organisms Transcription and translation can occur in genes transplanted from one species to another Transcription Defined as the DNA directed synthesis of RNA STEP 1 RNA polymerase separates the 2 DNA strands and connects RNA nucleotides as they base pair along the DNA template forms mRNA specifically premRNA Addition of RNA nucleotides 2. RNA polymerase add RNA only in the 3’ end, elongates in the 5’-3’ end Remember uracil replaces thymine First attachment is called the Promoter, and yes, it is called a TATA box which binds first to the DNA before RNA polymerase II can call transcription unit Terminator-end Synthesis of RNA transcript- stage 1Initiation Bacteria Eukaryotes RNA polymerase to the RNA polymerase II promoter cannot bind or attach without proteins called transcription factors- all called transcription initiation complex Crucial DNA Promoter is TATA that forms the complex RNA unwinds DNA Transcription Initiation Complex 2. Elongation of the RNA strand RNA polymerase moves along DNA Untwists helix RNA nucleotides added to 3’end Double helix reforms leaving hanging RNA strand 3. Termination After RNA polymerase hits a terminator sequence in DNA stops RNA transcript released Polymerase detaches Are we done yet? No!!! Modification of RNA after transcription ADDITION OF Poly-A tail 5’cap Helps with export from nucleus Protection from degradation of enzymes Attachment to ribosomes RNA Splicing of pre-mRNA Eukaryotic cells Introns are sections cut out Parts that are kept are exons Spliceosome do the splicing RNA or snRNA (small nuclear RNA) Ribozyme- RNA that acts as an enzyme Editing of premRNA intron and Exon Splicing What is a snRNPs? Particles called small nuclear ribonucleoproteins (called snurps) are small nuclear RNA plus a protein are called snRNP and these recognize splice sites Different snurps + additional protein form spliceosome Introns Have functional and evolutionary importance May regulate gene expression Genes can encode more than 1 kind of polypeptide depending on which segments are exons (alternative RNA splicing) Number of proteins (100,000)produced exceeds number of genes(25,000) Translation RNA directed synthesis of a polypeptide in the cytoplasm on the ribosome t-RNA Transfers correct t-RNA from pool in the cytoplasm R-RNA Accepts aa from t-RNA and binds aa at the other end with a triplet called anticodon Structure and function of tRNA Molecules of tRNA Not identical Each carries specific aa on one end Anticodon on one end base-pairs with the complementary codon on mRNA 80 nucleotides long Flattened into one plane, cloverleaf shape H bonds cause tRNA twist Roughly L-shaped Structure and Function of rRNA Complexes Proteins and tRNA 3 binding sites P-holds tRNA growing polypeptide chain A- holds tRNA that carries the amino acids added to next chain E-exit site for tRNA Ribosomes Facilitate coupling of tRNA anticodon to mRNA codons during protein synthesis Ribosomal subunits are made of proteins and ribosomal RNA (rRNA) Bacteria and Eukaryotic ribosomes are somewhat similar but have significant differences Antibiotic drugs specifically target bacterial ribosomes without harming eukaryotic ribosomes Figure 17.17 Growing polypeptide tRNA molecules E P Exit tunnel Large subunit A Small subunit 5 mRNA 3 (a) Computer model of functioning ribosome Growing polypeptide P site (Peptidyl-tRNA binding site) Exit tunnel Next amino acid to be added to polypeptide chain A site (AminoacyltRNA binding site) E site (Exit site) E mRNA binding site Amino end P A Large subunit Small subunit (b) Schematic model showing binding sites E tRNA mRNA 5 3 Codons (c) Schematic model with mRNA and tRNA 3 Stages of Translation Initiation Has 3 steps Elongation Has 3 steps Termination Stop codon in the mRNA is reached and translation stopped Initiation steps 1. mRNA with the code AUG is in the proper position 2. tRNA anticodon with UAC (methionine) hydrogen bonds to first with initiation factors assist in holding together 3. Subunit of ribosome allows methionine to attach to the P site Elongation Steps 1. Codon recognition Enters the A site Anticodon of incoming aminoacyl-tRNA base pairs withcomplement on A site GTP hydrolysis insures accuracy and efficiency Figure 17.16-1 Amino acid P P P Adenosine ATP Aminoacyl-tRNA synthetase (enzyme) Figure 17.16-2 Aminoacyl-tRNA synthetase (enzyme) Amino acid P Adenosine P P P Adenosine ATP P Pi Pi Pi Figure 17.16-3 Aminoacyl-tRNA synthetase (enzyme) Amino acid P Adenosine P P P Adenosine P Pi ATP Pi Pi tRNA Aminoacyl-tRNA synthetase tRNA Amino acid P Adenosine AMP Computer model Figure 17.16-4 Aminoacyl-tRNA synthetase (enzyme) Amino acid P Adenosine P P P Adenosine P Pi ATP Pi Pi tRNA Aminoacyl-tRNA synthetase tRNA Amino acid P Adenosine AMP Computer model Aminoacyl tRNA (“charged tRNA”) Elongation Steps 2. Peptide bond formation on P site rRNA catalyzes the formation of a peptide bond on the new amino acid to the carboxyl end of the growing polypeptide on the P site Removes the polypeptide from the A site to the P site Elongation Steps 3. Translocation Ribosomes translocates the tRNA in the A site to the P site, empty tRNA on the P site is moved to the E site where it is released Video of the Process http://www.hhmi.org/biointeractive/translation- basic-detail Termination A termination stop codon in the mRNA is reached Release factor (protein) binds to the stop codon Polypeptide is freed Wobble Is an mRNA triplet 64 different codons mRNA reads codon by codon, 1 aa added to the chain Relaxation of reading the 3rd base on t-RNA Accurate Translation 1. Correct match b/t tRNA and an amino acid Aminoacyl-tRNA synthetase (enzyme) Anticodon and mRNA codon match WOBBLE Flexible pairing at the 3rd base of a codon tRNA bind to more than one codon Completing and Targeting functional protein Translation is not Synthesis begins and enough Chains are modified Targeted to specific sites in the cell ends in cytosol unless the polypeptide signals the ribosome to attach to the ER Those destined for ER or secretion marked by signal peptide-Signal recognition particle (SRP) which brings the signal peptide to the ER Bound ribosomes Endomembrane-secreted out Free ribosomes-cytosol Protein folding Post-translational modifications Polyribosome Ribosomes can translate a single mRNA simultaneously Forms polyribosome or polysome Multiple copies of polypeptide quickly Figure 17.21 Growing polypeptides Completed polypeptide Incoming ribosomal subunits Start of mRNA (5 end) (a) End of mRNA (3 end) Ribosomes mRNA (b) 0.1 m Figure 17.21a Ribosomes mRNA 0.1 m Properties of RNA 3 properties of RNA 1.forms 3 dimensional structure can base-pair to itself 2. Some bases in RNA contain functional groups that may participate in catalysis 3. RNA can H bond to other nucleic acid molecules Figure 17.22 1 Ribosome 5 4 mRNA Signal peptide 3 SRP 2 ER LUMEN SRP receptor protein Translocation complex Signal peptide removed ER membrane Protein 6 CYTOSOL Mutations Affect protein structure and function Changes in the genetic material of the cell or virus Substitutions Nucleotide –pair substitution Silent mutation- no affect due to redundancy Missense-codes for incorrect aa Nonsense-codes to a stop codon and leads to nonfunctional Point mutations Single base Leads to the production of an abnormal protein Nucleotide –pair substitution Insertions and deletions Mutations Insertions and Delections Additions or losses of nucleotide pairs Disastrous effect on the resulting protein May alter the reading frame to a frameshift mutation Progeria Error LMNA gene, a protein that provides support to the cell nucleus. Accelerated aging Errors Insertions/deletions Mutagens Spontaneous mutations during DNA replication, recombination or repair Physical or chemical agents that cause mutations X-rays, UV light Aspertame Saccharin Polycyclic aromatic hydrocarbons- smoked meat and fish Bacteria, eukarya, Archaea Bacteria Eukarya Archaea Simultaneously transcribe Transcription and and translate the same translation are separated gene by the nuclear envelope Likely coupled of transcription and translation Proteins diffuse to sites Same size ribosomes as bacteria but function is different No transcription sitesaccessory proteins Resemble eukaryotes in RNA polymerase, termination of transcription and ribosomes Figure 17.26 TRANSCRIPTION DNA 3 5 RNA polymerase RNA transcript Exon RNA PROCESSING RNA transcript (pre-mRNA) AminoacyltRNA synthetase Intron NUCLEUS Amino acid AMINO ACID ACTIVATION tRNA CYTOPLASM mRNA Growing polypeptide 3 A Aminoacyl (charged) tRNA P E Ribosomal subunits TRANSLATION E A Anticodon Codon Ribosome