Molecular Biochemistry I
Electron Transfer Chain
Copyright © 1999-2007 by Joyce J. Diwan.
All rights reserved.
Electron Transfer
An electron transfer reaction:
Aox + Bred Ared + Box
Aox is the oxidized form of A (the oxidant)
Bred is the reduced form of B (the reductant).
For such an electron transfer, one may consider two
half-cell reactions:
Aox + n e- Ared
Box + n e- Bred
e.g., Fe+++ + e- Fe++
Aox + n e- Ared
Box + n e- Bred
For each half reaction:
E = E°' – RT/nF (ln [reduced]/[oxidized])
e.g., for the first half reaction:
E = E°' – RT/nF (ln [Ared]/[Aox])
E = voltage, R = gas const., F = Faraday, n = # of e-.
When [Ared] = [Aox],
E = E°'.
E°' is the mid-point potential, or standard redox
potential, the potential at which [oxidant] = [reductant]
for the half reaction.
For an electron transfer:
DE°' = E°'(oxidant) – E°'(reductant) = E°'(acceptor) – E°'(donor)
DGo' = – nFDE°'
(E°' is the mid-point potential)
An electron transfer reaction is spontaneous (negative
DG) if E°' of the donor is more negative than E°' of the
acceptor, i.e., when there is a positive DE°'.
Consider transfer of 2 electrons from NADH to oxygen:
a. ½ O2 + 2H+ + 2e- H2O
E°' = +0.815 V
b. NAD+ + 2H+ + 2e- NADH + H+
E°' = -0.315 V
Subtracting reaction b from a:
c. ½ O2 + NADH + H+ H2O + NAD+
DE°'= +1.13 V
DG = - nFDEo' = – 2(96494)(1.13) = – 218 kJ/mol
Electron Carriers
NAD+/NADH and FAD/FADH2 were introduced
earlier.
FMN (Flavin MonoNucleotide) is a prosthetic
group of some flavoproteins.
It is similar in structure to FAD (Flavin Adenine
Dinucleotide), but lacking the adenine nucleotide.
FMN (like FAD) can accept 2 e- + 2 H+ to form
FMNH2.
H
C
H3C
C
H3C
C
N
C
C
C
H
C
C
O H3C
C
N
CH2
-
HC
OH
HC
HC
FMN
H3C
NH
C
N
H2C
H
C
C
C
+
e +H
H
N
C
C
C
C
H
O H3C
C
-
+
e +H
H
N
C
C
C
C
H
C
NH
C
N
C
CH2
OH
OH
HC
OH
OH O
HC
OH O
OH O
HC
O
FMNH·
P
O-
O-
O
N
H
HC
HC
O-
C
CH2
H2C
C
N
OH
O-
H3C
NH
C
N
OH
P
H
C
C
HC
O
O
O
O
H2C
O
FMNH2
P
O-
O-
FMN, when bound at the active site of some enzymes, can
accept 1 e- to form the half-reduced semiquinone radical.
The semiquinone can accept a 2nd e- to yield FMNH2.
Since it can accept/donate 1 or 2 e-, FMN has an important
role mediating e- transfer between carriers that transfer 2e(e.g., NADH) & those that can accept only 1e- (e.g., Fe+++).
O
CH3O
CH3
CH3
CH3
CH3O
(CH2 CH
O
C
coenzyme Q
CH2)nH
H2C
C
C
CH2
H
isoprene
Coenzyme Q (CoQ, Q, ubiquinone) is very hydrophobic.
It dissolves in the hydrocarbon core of a membrane.
It includes a long isoprenoid tail, with multiple units
having a carbon skeleton comparable to that of isoprene.
In human cells, most often n = 10.
Q10’s isoprenoid tail is longer than the width of a bilayer.
It may be folded to yield a more compact structure, & is
postulated to reside in the central domain of a membrane,
between the 2 lipid monolayers.
O
CH3O
CH3
CH3
CH3O
(CH2 CH
O
C
CH2)nH
coenzyme Q
2 e- + 2 H+
OH
CH3O
The quinone ring of
coenzyme Q can be
reduced to the quinol
in a 2e- reaction:
CH3
CH3
CH3O
Q + 2 e- + 2 H+ QH2.
(CH2 CH
OH
C
CH2)nH
coenzyme QH2
O-
O
CH3O
CH 3
CH 3
CH3O
(CH 2 CH
O
C
e-
CH 2)nH
CH3O
CH 3
CH 3
CH3O
(CH 2 CH
O
coenzyme Q
C
CH 2)nH
coenzyme Q •-
e- + 2 H+
OH
CH3O
CH 3
CH 3
CH3O
(CH 2 CH
OH
C
CH 2)nH
coenzyme QH2
When bound to special sites in respiratory complexes,
CoQ can accept 1 e- to form a semiquinone radical (Q·-).
Thus CoQ, like FMN, can mediate between 1 e- & 2 edonors/acceptors.
Coenzyme Q functions as a mobile e- carrier within
the mitochondrial inner membrane.
Its role in trans-membrane H+ transport coupled to
e- transfer (Q Cycle) will be discussed later.
CH3
CH3
S
HC
CH2 protein
N
H3C
CH3
N
-
OOC
CH2 CH2
Fe
N
CH
N
S
CH2
protein
CH3
CH2
CH3
CH2
COO-
Heme c
Heme is a prosthetic group of cytochromes.
Heme contains an iron atom in a porphyrin ring system.
The Fe is bonded to 4 N atoms of the porphyrin ring.
CH3
CH3
S
HC
CH2 protein
N
H3C
CH3
N
-
OOC
CH2 CH2
Fe
N
CH
N
S
CH2
protein
CH3
CH2
CH3
CH2
COO-
Heme c
Hemes in the 3 classes of cytochrome (a, b, c) differ slightly in
substituents on the porphyrin ring system. A common feature is
2 propionate side-chains. Only heme c is covalently linked to
the protein via thioether bonds to cysteine residues.
CH3
CH2
CH3
HC
CH2
CH
C
CH2
3
H
OH
O
N
HC
CH3
N
-
OOC
Fe
CH2 CH2
N
CH
N
CH2
CH2
CH3
CH2
COO-
Heme a
Heme a is unique in having a long farnesyl side-chain
that includes 3 isoprenoid units.
PDB file 5CYT
In the RasMol display of
heme c at right, the
porphyrin ring system is
displayed as ball & sticks,
while Fe is displayed as
spacefill.
Heme in cytochrome c
The heme iron can undergo a 1 e- transition between
ferric and ferrous states: Fe+++ + e- Fe++
The porphyrin ring is planar.
The heme Fe is usually bonded
to 2 axial ligands, above &
below the heme plane (X,Y) in
addition to 4 N of porphyrin.
PDB file 5CYT
His
Met
Heme in cytochrome c
X
N
N
Fe
N
N
Y
Axial ligands may be S or N
atoms of amino acid side-chains.
Axial ligands in cyt c are Met S
(yellow) and His N (blue).
A heme that binds O2 may have
an open (empty) axial ligand
position.
Cytochromes are proteins with heme prosthetic groups.
They absorb light at characteristic wavelengths.
Absorbance changes upon oxidation/reduction of the
heme iron provide a basis for monitoring heme redox state.
Some cytochromes are part of large integral membrane
complexes, each consisting of several polypeptides &
including multiple electron carriers.
Individual heme prosthetic groups may be separately
designated as cytochromes, even if in the same protein.
E.g., hemes a & a3 that are part of the respiratory chain
complex IV are often referred to as cytochromes a & a3.
Cytochrome c is instead a small, water-soluble protein
with a single heme group.
Cytochrome c
PDB
5CYT
heme
complex IV
cyt. c
Lys13
Lys 72
Positively charged lysine residues (in magenta) surround
the heme crevice on the surface of cytochrome c.
These may interact with anionic residues on membrane
complexes to which cyt c binds, when receiving or
donating an e-.
Cys
S
Fe
S
S
Cys
Cys
Fe
Fe
S
S
S
Cys
S
S
Fe
Cys
S
S
Fe
Cys
S
S
Cys
S
Cys
Fe
S
Iron-Sulfur Centers
Fe-S
spacefill;
cysteine ball
& stick.
Fe orange;
2-iron Fe-S S yellow.
4-iron Fe-S
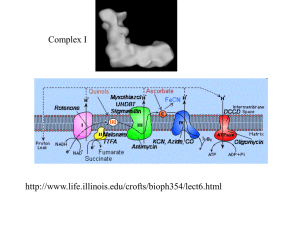
Two iron-sulfur centers
from complex I
PDB 2FUG
Iron-sulfur centers (Fe-S) are prosthetic groups containing
2, 3 , 4 or 8 iron atoms complexed to elemental & cysteine S.
4-Fe centers have a tetrahedral structure, with Fe & S atoms
alternating as vertices of a cube.
Cysteine residues provide S ligands to the iron, while also
holding these prosthetic groups in place within the protein.
Cys
Fe
S
Electron transfer proteins may
contain multiple Fe-S centers.
Iron-sulfur centers transfer
only one electron, even if they
contain two or more iron
atoms, because of the close
proximity of the iron atoms.
S
Cys
Cys
Fe
S
Fe
S
S
S
Cys
S
S
Fe
Cys
S
S
Fe
Cys
S
S
Cys
S
Cys
Fe
S
Iron-Sulfur Centers
E.g., a 4-Fe center might cycle between redox states:
Fe+++3, Fe++1 (oxidized) + 1 e- Fe+++2, Fe++2 (reduced)
matrix
cristae
Respiratory
Chain:
intermembrane
space
inner
membrane mitochondrion
outer
membrane
Most constitutents of the respiratory chain are
embedded in the inner mitochondrial membrane (or
in the cytoplasmic membrane of aerobic bacteria).
The inner mitochondrial membrane has infoldings
called cristae that increase the membrane area.
Electrons are
Matrix
transferred from H+ + NADH NAD+ + 2H+
NADH O2 via
2
e
multisubunit
I
Q
III
inner membrane
complexes I, III
& IV, plus CoQ Intermembrane Space
& cyt c.
2H+ + ½ O2 H2O
IV
cyt c
Within each complex, electrons pass sequentially through
a series of electron carriers.
CoQ is located in the lipid core of the membrane. There
are also binding sites for CoQ within protein complexes
with which it interacts.
Cytochrome c resides in the intermembrane space.
It alternately binds to complex III or IV during e- transfer.
Individual
respiratory
chain complexes
have been
isolated and their
composition
determined.
Matrix
H+ + NADH NAD+ + 2H+
I
2 eQ
2H+ + ½ O2 H2O
III
Intermembrane Space
IV
cyt c
There is also evidence for the existence of stable
supramolecular aggregates containing multiple
complexes.
E.g., complex I, which transfers electrons to coenzyme Q,
may associate with complex III, which reoxidizes the
reduced coenzyme Q, to provide a pathway for direct
transfer of coenzyme Q between them.
Composition of Respiratory Chain Complexes
No. of
Proteins
Prosthetic
Groups
Complex
Name
Complex I
NADH
Dehydrogenase
46
FMN,
9 Fe-S cntrs.
Complex II
Succinate-CoQ
Reductase
5
FAD, cyt b560,
3 Fe-S cntrs.
Complex III
CoQ-cyt c
Reductase
11
cyt bH, cyt bL,
cyt c1, Fe-SRieske
Complex IV
Cytochrome
Oxidase
13
cyt a, cyt a3,
CuA, CuB
Matrix
H+ + NADH NAD+ + 2H+
I
2 eQ
2H+ + ½ O2 H2O
III
Intermembrane Space
IV
cyt c
Mid-point potentials of constituent e- carriers are
consistent with the e- transfers shown being spontaneous.
Respiratory chain inhibitors include:
Rotenone (a rat poison) blocks complex I.
Antimycin A blocks electron transfer in complex III.
CN- & CO inhibit complex IV.
Inhibition at any of these sites will block e- transfer from
NADH to O2.
NAD+
Complex I
catalyzes
oxidation of
NADH, with
reduction of
coenzyme Q:
NADH
FMN
peripheral
domain
matrix
inner mitochondrial
membrane
membrane domain
Complex I
NADH + H+ + Q NAD+ + QH2
Transmembrane H+ flux associated with this reaction will
be discussed in the section on oxidative phosphorylation.
An atomic-level structure is not yet available for the entire
complex I, which in mammals includes at least 46 proteins,
along with prosthetic groups FMN & several Fe-S centers.
NAD+
NADH
FMN
peripheral
domain
matrix
Complex I is
L-shaped.
inner mitochondrial
membrane
membrane domain
Complex I
The peripheral domain, containing the FMN that accepts
2e- from NADH, protrudes into the mitochondrial matrix.
Iron-sulfur centers are also located in the hydrophilic
peripheral domain, where they form a pathway for etransfer from FMN to coenzyme Q.
A binding site for coenzyme Q is thought be close to the
interface between peripheral and intra-membrane domains.
The initial electron transfers are:
NADH + H+ + FMN NAD+ + FMNH2
FMNH2 + (Fe-S)ox FMNH· + (Fe-S)red + H+
After Fe-S is reoxidized by transfer of the electron to
the next iron-sulfur center in the pathway:
FMNH· + (Fe-S)ox FMN + (Fe-S)red + H+
Electrons pass through a series of iron-sulfur centers,
and are eventually transferred to coenzyme Q.
Coenzyme Q accepts 2 e- and picks up 2 H+ to yield
the fully reduced QH2.
NAD+
NADH
An X-ray structure
has been determined
for the hydrophilic
peripheral domain of
a bacterial complex I
FMN
peripheral
domain
matrix
inner mitochondrial
membrane
membrane domain
Complex I
This bacterial complex I contains fewer proteins than the
mammalian complex I, but includes the central subunits
found in all prokaryotic & eukaryotic versions of complex I.
The prosthetic groups are found to be all in the
peripheral domain, that in the mammalian complex would
protrude into the mitochondrial matrix.
Peripheral domain of a bacterial Complex I
A
FMN
Iron-sulfur centers
are arranged as a
wire, providing a
pathway for etransfer from FMN
through the protein.
B
FMN
membrane
domain
N2
PDB 2FUG
N2, the last Fe-S center in the chain, passes e- one at a
time to the mobile lipid redox carrier coenzyme Q.
A proposed binding site for CoQ is close to N2 at the
interface of peripheral & membrane domains.
P. L. Dutton and coworkers have called attention to the
relevance of conserved distances between redox carriers
within respiratory chain complexes with regard to the
energy barrier at each step for electron tunneling
through the protein.
They have modeled electron transfers through the
respiratory chain complexes, and provide an animation
of the time course of electron transfer through Complex I.
For more diagrams see
A review by U. Brandt (requires Annual Reviews
subscription).
The Complex I Home Page
COO-
Succinate Dehydrogenase
of the Krebs Cycle is also
called complex II or
Succinate-CoQ Reductase.
FAD is the initial eacceptor.
H
C
H
H
C
H
Q
QH2
COOC
via FAD H
COO-
H
C
COO-
succinate
fumarate
Succinate Dehydrogenase
(Complex II)
FAD is reduced to FADH2 during oxidation of succinate
to fumarate.
FADH2 is then reoxidized by transfer of electrons through
a series of 3 iron-sulfur centers to CoQ, yielding QH2.
The QH2 product may be reoxidized via complex III,
providing a pathway for transfer of electrons from
succinate into the respiratory chain.
Complex II
OAA
X-ray crystallographic
analysis of E. coli
complex II indicates a
linear arrangement of
electron carriers within
complex II, consistent with
the predicted sequence of
electron transfers:
FAD
FeS
membrane
domain
CoQ
heme
PDB 1NEK
FAD FeScenter 1 FeScenter 2 FeScenter 3 CoQ
In this crystal structure oxaloacetate (OAA) is bound in
place of succinate.
Matrix
H+ + NADH NAD+ + 2H+
I
2 eQ
2H+ + ½ O2 H2O
III
Intermembrane Space
IV
cyt c
Complex III accepts electrons from coenzyme QH2 that
is generated by electron transfer in complexes I & II.
The structure and roles of complex III are discussed in
the class on oxidative phosphorylation.
Cytochrome c1, a prosthetic group within complex III,
reduces cytochrome c, which is the electron donor to
complex IV.
Matrix
H+ + NADH NAD+ + 2H+
I
2 eQ
2H+ + ½ O2 H2O
III
Intermembrane Space
IV
cyt c
Cytochrome oxidase (complex IV) carries out the
following irreversible reaction:
O2 + 4 H+ + 4 e- 2 H2O
The four electrons are transferred into the complex one
at a time from cytochrome c.
membrane
Cytochrome oxidase dimer
(PDB file 1OCC)
Intramembrane domains of cytochrome oxidase
(complex IV) consist mainly of transmembrane
a-helices.
Complex IV
binuclear center
heme a3
CuB
PDB 1OCC
Metal centers of cytochrome oxidase (complex IV):
heme a & heme a3,
CuA (2 adjacent Cu atoms) & CuB.
O2 reacts at a binuclear center consisting of heme a3
and CuB.
PDB file 1OCC
Metal center ligands in
complex IV:
Heme axial ligands are His N
atoms.
Heme a is held in place
between 2 transmembrane
a-helices by its axial His
ligands.
Liganding of Heme a
in Cytochrome Oxidase
Heme a3, which sits adjacent to
CuB, has only one axial ligand.
Cu ligands consist of His N, & in
the case of CuA also Cys S, Met S,
& a Glu backbone O.
binuclear center
His ligands
heme a3
Electrons enter complex IV one at
a time from cyt c to CuA.
They then pass via heme a to the
binuclear center where the
chemical reaction takes place.
CuB
PDB 1OCC
Electron transfers: cyt c → CuA → heme a → heme a3/CuB
O2 binds at the open axial ligand position of heme a3,
adjacent to CuB.
binuclear center
His ligands
O2 + 4 H+ + 4 e- 2 H2O
Details of the reaction sequence
are still debated.
heme a3
A Tyr-His complex adjacent to the
binuclear center is postulated to
have a role in O-O bond splitting.
CuB
PDB 1OCC
The open axial ligand position makes heme a3 susceptible
to binding each of the following inhibitors:
CN-, CO, and the radical signal molecule ·NO.
·NO may regulate cell respiration through its inhibitory
effect, & can induce a condition comparable to hypoxia.