BIOLUMINESCENT SENSORS
advertisement

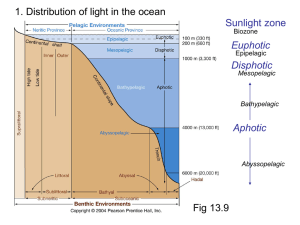
BIOLUMINESCENT SENSORS JING WANG Department of Nutrition and Food Science ENPM808B Dec 3rd, 2003 Outline Structure of Biosensor Bioluminescent bacteria Target Analytes Transducers Applications Summary Structure of Biosensor Analyte BioReceptor Transducer Measurable Signal Bioluminescent bacteria Firefly Bioluminescence firefly luciferase oxyluciferin + PPi + CO2 + h luciferin + ATP + O2 Mg2+ (max = 560 nm) Bacterial Bioluminescence Vibrio Photobacterium Photobacterium phosphoreum Xenorhabdus Xenorhabdus nematophilus Bioluminescence luxCDABE Genes synthetase RCOOH + ATP + NADPH NADP + AMP + Ppi + RCHO reductase luciferase transferase RCOX + HOH(HSR’) RCOOH(RCOSR’) + XH FMNH2 + RCHO + O2 FMN +H2O + RCOOH +h(490nm) Figure 1. Bacterial bioluminescence pathway (adapted from Van Dyk, 1998) Figure 2. cloning of bioluminescent gene into E. coli strains Target Analytes Inorganic Substances Mercury Hg Potassium nitrate KNO3 Nickel Ni Organic Substances Phenol Octane Benzene Ethanol Urea Naphthalene Transducer Photomultipier Tubes Luminometer Turner BioSystems' TD-20/20 single-tube luminometer Applications Light out Quantitating loss of bioluminescence due to the toxicity of the sample tested or of the environmental condition imposed Light on The choice of the promoter driving expression of the lux genes determines the specificity of the response Example 1 Monitoring and classification of PAH toxicity using an immobilized bioluminescent bacteria Hyun Joo Lee, Julien Villaume, David C. Cullen, Byoung Chan Kim, Man Bock Gu. Biosensors and Bioelectronics, Volume 18, Issues 5-6, May 2003, Pages 571-577 Background Polycyclic Aromatic Hydrocarbons (PAHs) PAHs are a class of very stable organic molecules made up of only carbon and hydrogen. These molecules are flat, with each carbon having three neighboring atoms much like graphite. CCPAHs Naphthalene Phenanthrene Anthracene PCPAHs Pyrene Benzo[a]pyrene Materials and methods Recombinant E. Coli Strain RFM443 Immobilization Procedure Materials and methods Ampicillin 100g/ml Centrifuge 6000rpm 50 ml sample E. Coli GC2 cells Polypropylene tubes 10 min 500 l fresh 25 ºC LB medium Sterile glass beads (0.05 g, 150 to 212 m) 20 ml Agar Media Collected Cells 10 mm 100 l cell mixture Materials and methods Recombinant E. Coli Strain RFM443 Solubilization of PAHs Using Rhamnolipids as Biosurfactant Immobilization Procedure Measurement System Materials and methods Schematic diagram of the soil biosensor system Results and Discussion Relative Bioluminescence (RBL) The ratio of the test bioluminescence to the control’s bioluminescence Results and Discussion Bioluminescent response to PCPAHs (a) pyrene (b) benzo[a]pyrene Results and Discussion Bioluminescent response to CCPAHs (a) naphthalene (b) anthracene Results and Discussion Bioluminescent response to CCPAHs (c) phenanthrene Conclusions The response patterns of this soil biosensor system to CCPAHs or PCPAHs were clearly identifiable. Only CCPAHs were found to cause toxicity and inhibit cellular metabolism, while PCPAHs did not affect any changes in bioluminescence responses. Example 2 Construction and characterization of novel dual stress-responsive bacterial biosensors Robert J. Mitchell and Man Bock Gu. Biosensors and Bioelectronics, In Press, Corrected Proof, Available online 18 November 2003 Background Green Fluorescence Protein (GFP) Xenorhabdus luminescens (Photorhabdus luminescens) Materials and Methods two stress-responsive Escherichia coli biosensor strains Divergent Orientation Tandem Orientation Figure 3. Fusion gene constructs used in this study Materials and Methods Hydroxyl radical-forming chemicals Hydrogen Peroxide Cadmium Chloride Materials and Methods Genotoxins Mitomycin C Methyl-N-nitro-Nnitrosoguanidine (MNNG) Materials and Methods General toxincants Isopropanol Ethanol CH3CH2OH Phenol 100 l 100 l chemical opaque Plate luminometer 250 ml flask 50 ml LB medium 100 l chemical E. coli strains clear 100 l 96-well plate FLx800 Microplate fluorometer Results and Discussion Figure 4. Time-dependent plots of the fluorescent response from DUO-1 after exposure to various concentrations of (a) mitomycin C and (b) MNNG Table1. Response characteristics of DUO-1 and DUO-2 a Concentration (mg/l) giving the maximum induction b NR: no response; RBL or FL value of less than 2.0 and 1.25, respectively c Value in parenthesis is the lowest concentration (mg/l) giving a twofold induction of bioluminescence or a maximum slope of 0.01 Figure 5. Time-dependent bioluminescent plots from DUO-1 (a and c) and DUO-2 (b and d) after exposure to various concentrations of hydrogen peroxide (a and b) and mitomycin C (c and d) Conclusions Both strains showed an induction of green fluorescent protein (GFP) and bioluminescence when they experienced DNA and oxidative damage, respectively. Conclusions The tandem orientation of the two fusion genes within DUO-2 allowed it to sensitively respond to genotoxins via the production of bioluminescence. The characteristics of DUO-2's bioluminescent response to each stress were easily distinguishable, making it useful for the detection of both stresses. Conclusions Furthermore, tests with mixtures of chemicals showed that both DUO-1 and DUO-2 were responsive when chemicals causing oxidative or genotoxic stress were present as a single chemical or within complex chemical mixtures. Summary Advantages Quick response time Not sensitive to environmental changes Easy to operate and control Disadvantages Difficult to remain the cell alive and viable Not very stable during the sensing time Less specific comparing to other types of biosensors