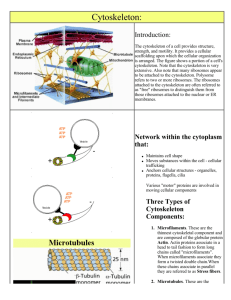
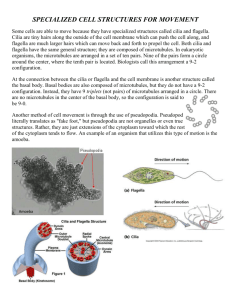
Cytoskeleton
advertisement

A. Overview B. Experimental Methods C. Microtubules D. Microfilaments Cytoskeleton A. B. C. D. Overview Experimental Methods Microtubules Microfilaments (Updated 4/9/08) A. Overview B. Experimental Methods C. Microtubules D. Microfilaments A. Overview 1. 2. 3. 4. Definition Types of Cytoskeleton Fibers Dynamic Polymerization/Depolymerization Molecular Motors Alberts: Fig. 16 – 1, Panel 16 – 1, Panel 16 – 2, Fig. 16 –11, Fig 16 – 12, 16 – 8, 16 – 7, 16 – 10, 16 – 13, 16 – 14, 16 – 15, 16 – 16, 16 – 17, 16 – 19, 16 – 56, Table 16 – 1 A. Overview B. Experimental Methods C. Microtubules D. Microfilaments B. Experimental methods 1. Visualization Approaches – – Light Microscopy Fluorescence Microscopy » – – 2. 3. http://www.itg.uiuc.edu/exhibits/gallery/fluorescencegallery.htm Digital/video Microscopy Electron Microscopy Genetic Approaches Biochemical Approaches A. Overview B. Experimental Methods C. Microtubules 1. Structure 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs C. Microtubules 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments 1. 2. 3. 4. 5. 6. 7. Structure Microtubule-associated proteins Functions Microtubule motors Microtubule organizing centers Dynamic properties of microtubules Flagella and cilia A. Overview B. Experimental Methods C. Microtubules 1. Structure C.1. Microtubules: Structure 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments 1. Structure – – Alberts: Fig 16 – 11 Structure and composition - hollow, tubular; found in most eukaryotic cells (cilia, spindle, flagella) » » » – – – Outer diameter - 24 nm Wall thickness - ~5 nm May extend across cell length/breadth Wall composed of globular proteins arranged in longitudinal rows (protofilaments) Protofilaments are aligned parallel to tubule long axis In cross section, consist of 13 protofilaments arrayed in circular pattern within wall A. Overview B. Experimental Methods C. Microtubules 1. Structure 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs C.1. Microtubules: Structure 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments – Each protofilament is assembled of dimeric building blocks (one a-tubulin & one b-tubulin; A heterodimer) organized in linear array along length of protofilament – Two types of tubulin subunits have similar 3D structure & fit tightly together – Protofilaments asymmetric (a-tubulin at one end, btubulin at other); All in single MT have same polarity; Each assembly unit has 2 nonidentical components (heterodimer) – All protofilaments of microtubule have same polarity; Thus so does full tubule (plus- & minus-end) – Plus end - fast-growing (b-tubulins on tip); Minus end slow-growing (a-tubulins on tip) A. Overview B. Experimental Methods C. Microtubules 1. Structure 2. MAPs C.2. Microtubules: MAPs 3. Functions 4. Microtubule Motors 5. MTOCs 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments 2. Microtubule-associated proteins – – – – Alberts: Fig 16-40, 16-41 MTs can assemble in vitro from purified tubulin, but MAPs are found with MTs isolated from cells; most found only in brain tissue; MAP4 has wider distribution Have globular head domain that attaches to MT side & filamentous tail protruding from MT surface May interconnect MTs to help form bundles (crossbridges), increase MT stability, alter MT rigidity, influence MT assembly rate A. Overview B. Experimental Methods C. Microtubules 1. Structure 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs C. 2. Microtubules: MAPs 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments – MAP activity controlled by addition & removal of phosphate groups from particular amino acid residues by protein kinases & phosphatases, respectively; example - Alzheimer’s disease (AD) – Abnormally high MAP (tau) phosphorylation implicated in fatal neurodegenerative diseases like AD; neurofibrillary tangles in brains made of hyperphosphorylated tau; may help kill nerve cells – Excessively phosphorylated tau molecules are unable to bind to MTs; people with one of these diseases, a type of dementia called FTDP-17, carry mutations in tau gene, implicating it as cause A. Overview B. Experimental Methods C. Microtubules 1. Structure 2. MAPs 3. Functions C. 3. Microtubules: Functions 4. Microtubule Motors 5. MTOCs 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments 3. Functions – Alberts: Table 16-2; Fig 16-23, 66 Internal skeleton (scaffold) providing structural support & maintaining organelle position – Resist compression or bending forces on fiber; provide mechanical support like girders in `tall building; prevent distortion of cell by cytoplasmic contractions – MT distribution conforms to & helps determine cell shape: flattened, round cells - radiate from nuclear area; columnar epithelium - parallel to cell long axis; like aluminum rods support tent A. Overview B. Experimental Methods C. Microtubules 1. Structure 2. MAPs 3. Functions C. 3. Microtubules: Functions 4. Microtubule Motors 5. MTOCs 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments – Elongated cell process (axon, axopods of heliozoan protists) - MTs oriented parallel to each other & axon or axopod long axis; help move things – In developing embryo, extend growing central NS axons to peripheral NS; inhibit (colchicine [CO], nocodazole [NO]) & outgrowth stops, regresses (collapses back to rounded cell body) – Found as core of axopodial processes of heliozoan protozoa; many MTs arranged in spiral with individual MTs traversing entire length of process A. Overview B. Experimental Methods C. Microtubules 1. Structure 2. MAPs 3. Functions C. 3. Microtubules: Functions 4. Microtubule Motors 5. MTOCs 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments Plants: play similar role in plants; affect shape indirectly by influencing cell wall formation; found in cortex just below membrane during interphase forming a distinct cortical zone A. Overview B. Experimental Methods C. Microtubules 1. Structure C. 3. Microtubules: Functions 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments Also have role in maintenance of cell internal organization (organelle placement) - disrupt MTs (CO, NO) —> Golgi disperses to cell periphery; goes back to cell center when inhibitors removed Move macromolecules & organelles around cell in directed manner (intracellular motility) – – Halt vesicle transport between compartments if disrupt MTs so transport dependent on them Proteins made in neuron cell body move down axon (neurotransmitters, etc.) in vesicles A. Overview B. Experimental Methods C. Microtubules 1. Structure C. 3. Microtubules: Functions 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs 6. Dynamic Properties – Different materials move at different rates; fastest rate is 5 µm/sec (400 mm/day); vesicles seen attached to MTs – Structures & materials moving toward neuron terminals are said to move anterograde – Other structures, like endocytic vesicles that are formed at neuron terminals & carry regulatory factors from target cells, move from synapse to cell body in a retrograde direction – Ex.: axons filled with MTs, MFs & IFs; evidence suggests that both anterograde & retrograde movement are mediated mostly by MTs; video microscopy shows vesicles moving along MTs – Confirmed by EM of axons; molecular motors move vesicles along the MTs that serve as tracks 7. Flagella and Cilia D. Microfilaments A. Overview B. Experimental Methods C. Microtubules 1. Structure C. 3. Microtubules: Functions 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments Motile elements of cilia & flagella (more later) Active components of mitotic/meiotic machinery; move chromosomes A. Overview B. Experimental Methods C. Microtubules 1. Structure 2. MAPs C. 4. Microtubules: Motors 3. Functions 4. Microtubule Motors 5. MTOCs 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments 4. Microtubule motors – Alberts: Fig 16-58, 59, 60, 62, 63, 64, 67 Motor proteins: convert chemical energy stored in ATP into mechanical energy that is used to move cellular cargo attached to motor – – Types of cellular cargo transported by these molecular motors include: vesicles, organelles (mitochondria, lysosomes, chloroplasts), chromosomes, other cytoskeletal filaments A single cell may contain dozens of different motor proteins, each specialized for moving a particular type of cargo in particular cell region A. Overview B. Experimental Methods C. Microtubules 1. Structure 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs C. 4. Microtubules: Motors 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments – Collectively, motor proteins are grouped into 3 broad families: myosins, kinesins, dyneins » Kinesins & dyneins move along MTs; myosins move along MFs; None known for ifs » Motor proteins move unidirectionally along their cytoskeletal track in a stepwise manner from one binding site to the next » As they move along, they undergo a series of conformational changes (a mechanical cycle) » Steps of mechanical cycle are coupled to chemical cycle, which provides energy fueling movement » Includes motor binding ATP, ATP hydrolysis, product (ADP & Pi) release & binding of new ATP » Binding & hydrolysis of 1 ATP moves motor a few nm along track; Cycles repeated many times A. Overview B. Experimental Methods C. Microtubules 1. Structure 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs C. 4. Microtubules: Motors 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments Kinesins – Kinesins move vesicles/organelles from cell body to synaptic knobs; isolated in 1985 from squid giant axons; tetramer made of 2 identical heavy chains & 2 identical light chains; smallest & best understood – Large protein - pair of globular heads generate force by hydrolyzing ATP & bind MT; each head connected to a neck, a rodlike stalk & fan-shaped tail that binds cargo to be hauled – Diverse superfamily of kinesins - heads similar since roles similar; tails vary since they haul different cargoes A. Overview B. Experimental Methods C. Microtubules 1. Structure 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs C. 4. Microtubules: Motors 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments – In vitro mobility assay - kinesin-coated beads move to MT "+" end (axon tip); it is a "+" end-directed MT motor, therefore, kinesin responsible for anterograde movement » All MTs of axon are oriented with"-" ends facing cell body & "+" ends facing synaptic knobs » Moves through ATP-dependent cross-bridge cycle along single MT protofilament (rate proportional to [ATP]; up to ~1 µm/sec); at low concentrations, move slowly & see movement in distinct steps » Each step is ~8 nm in length, the spacing between tubulin dimers along protofilament » Appear to move 2 globular subunits (or 1 dimer at a time); usually toward membrane & "+" ends A. Overview B. Experimental Methods C. Microtubules 1. Structure 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs C. 4. Microtubules: Motors 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments – Kinesin possesses 2 motor domains that work by “hand-over-hand” mechanism; one always firmly attached to MT » 2 heads of kinesin behave in coordinated manner, so that they are always present at different stages in their chemical & mechanical cycles at a given time » When one head binds to MT, the interaction induces a conformational change in adjacent neck region of motor protein; it swings the other head forward toward binding site on next dimer » Force generated by head catalytic activity leads to swinging movement of neck » A kinesin molecule walks along a MT, hydrolyzing one ATP with each step A. Overview B. Experimental Methods C. Microtubules 1. Structure 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs C. 4. Microtubules: Motors 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments Conventional kinesin (discovered in 1985) is only one member of a superfamily of related kinesins – Mammalian genome sequence analysis leads to estimate that mammals make >50 different kinesins – Heads of kinesins have related amino acid sequences, reflecting common evolutionary ancestry & their similar role in moving along MTs – In contrast, kinesin tails have diverse sequences, reflecting variety of cargo different proteins haul A. Overview B. Experimental Methods C. Microtubules 1. Structure 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs C. 4. Microtubules: Motors 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments – Most kinesins travel toward the "+" end; but one small subfamily of kinesins (including the Drosophila Ncd protein) moves toward the MT "-" end » one would expect that the heads of "+"- & "-"-directed would have a different structure since the heads contain the catalytic core of the motor domain » But the heads are virtually indistinguishable; instead differences in direction of movement are determined by differences in the adjacent neck regions of the two proteins » When the head of a "-" end-directed Ncd molecule is joined to the neck-stalk portions of a kinesin molecule, the hybrid protein moves toward the "+" end of a MT track » Even if the hybrid has a catalytic domain that would normally move toward the "-" end of a MT, as long as it is joined to the neck of a "+" end motor, it moves in the "+" direction A. Overview B. Experimental Methods C. Microtubules 1. Structure 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs C. 4. Microtubules: Motors 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments A third subfamily of kinesinlike proteins is incapable of movement: kinesins of this group, like KXKCM1, are thought to destabilize MTs rather than acting as MT motors A. Overview B. Experimental Methods C. Microtubules 1. Structure 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs C. 4. Microtubules: Motors 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments Cytoplasmic Dyneins – Dyneins - first MT-associated motor found (1963); responsible for moving cilia & flagella – Thought to be ubiquitous eukaryotic motor protein; related protein found in variety of nonneural cells – Cilia/flagella form of protein was called axonemal dynein; its new relatives were called cytoplasmic dynein – Huge protein (~1.5 million daltons); 2 identical heavy chains & variety of intermediate & light chains – Each dynein heavy chain forms large globular head (~10X larger than a kinesin head) that generates force; moves along MT toward "-" end A. Overview B. Experimental Methods C. Microtubules 1. Structure 2. MAPs C. 4. Microtubules: Motors 3. Functions 4. Microtubule Motors 5. MTOCs 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments Suggested roles of cytoplasmic dynein – – – – Force generating agent for chromosome movement in mitosis "-"-directed MT motor for Golgi complex positioning & movement of vesicles/organelles through cytoplasm In nerve cells, cytoplasmic dynein involved in axonal retrograde organelle movement (toward cell body & cell center) & anterograde movement of MTs Fibroblasts & other nonneural cells: may move varied membranous organelles (endosomes, lysosomes, ERderived vesicles going toward Golgi) from periphery toward cell center A. Overview B. Experimental Methods C. Microtubules 1. Structure 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs C. 4. Microtubules: Motors 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments – Cytoplasmic dynein does not interact directly with membrane-bounded cargo, but requires intervening multisubunit complex, dynactin that may regulate dynein activity & help bind it to MT – Present model may be overly simplistic: kinesin & cytoplasmic dynein move similar materials in opposite directions over the same railway network – Individual organelles may bind kinesin & dynein simultaneously although only one is active at given time; myosin may also be present on some of these organelles A. Overview B. Experimental Methods C. Microtubules 1. Structure 2. MAPs C. 5. Microtubules: MTOCS 3. Functions 4. Microtubule Motors 5. MTOCs 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments 5. Microtubule-organizing centers – – Alberts: Panel 16-1; Fig 16-29, 30, 31, 32, 33 Function of MT in living cell depends on its location & orientation, thus it is important to understand why a MT assembles in one place as opposed to another controlled by MT-organizing centers (MTOCs) A. Overview B. Experimental Methods C. Microtubules 1. Structure 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs C. 5. Microtubules : MTOCS 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments Assembly of MTs from ab-dimers occurs in 2 distinct phases – Nucleation - slower; small portion of MT initially formed; occurs in association with specialized structures in vivo called microtubule-organizing centers (MTOCs); centrosome is example – Elongation - more rapid A. Overview B. Experimental Methods C. Microtubules 1. Structure 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs C. 5. Microtubules : MTOCS 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments Centrosomes - complex structure with 2 barrelshaped centrioles surrounded by amorphous, electron dense pericentriolar material (PCM) – In animal cells, cytoskeleton MTs typically form in association with centrosome – Centrioles: cylindrical; ~0.2 nm dia & typically ~twice as long; usually with 9 evenly spaced fibrils – Each fibril seen in cross section to be composed of 3 fused MTs (A, [the only complete one]; B & C), A is attached to centriole center by radial spoke A. Overview B. Experimental Methods C. Microtubules 1. Structure 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs C. 5. Microtubules : MTOCS 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments – 3 MTs of each triplet arranged in pattern that gives centriole a characteristic pinwheel appearance – Centrioles usually in pairs at right angles to each other near cell center just outside nucleus – Extraction of isolated centrosomes with 1 M potassium iodide removes ~90% of PCM protein leaving behind spaghetti-like scaffold of insoluble fibers – Centrosomes are sites of convergence of large numbers of MTs A. Overview B. Experimental Methods C. Microtubules 1. Structure 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs C. 5. Microtubules : MTOCS 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments MT polymerization & disassembly - treat with poisons (CO, NO) or cold —> MTs disassemble; much has been learned about their disassembly & reassembly in cultured animal cells in this way – Observe assembly when cells warmed or poisons removed; fix at various times after & stain with fluorescent anti-tubulin ABs – Within a few minutes of inhibition removal, 1 or 2 bright fluorescent spots seen in cytoplasm – Within 15 - 30 minutes, number of labeled filaments radiating from these foci rises dramatically A. Overview B. Experimental Methods C. Microtubules 1. Structure 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs C. 5. Microtubules : MTOCS 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments – In EM: MTs radiate out from centrosome; MTs don't actually penetrate into centrosome & contact centrioles, but terminate in dense pericentriolar material residing at centrosome periphery – PCM apparently initiates MT formation; centrioles not involved in MT nucleation, but they probably play a role in recruiting surrounding PCM during centrosome assembly A. Overview B. Experimental Methods C. Microtubules 1. Structure C. 5. Microtubules : MTOCS 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments Centrosome typically situated near center of cell, just outside nucleus – Columnar epithelium - centrosome moves from cell center to apical region just beneath cortex; cytoskeletal MTs emanate from site, extending toward nucleus & basal surface of cell – Regardless of location, centrosomes are sites of MT nucleation; polarity is always the same: "-" end at centrosome, "+" (growing) end at opposite tip – Thus, even though MTs are nucleated at MTOC, they are elongated at opposite end of polymer A. Overview B. Experimental Methods C. Microtubules 1. Structure 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs C. 5. Microtubules : MTOCS 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments Not all MTs associated with centrosome; – some animal cells (mouse oocytes) lack centrosomes entirely, but still make spindle – MTs of axon are not associated with centrosome, which is located in cell body, but they may be formed at centrosome, then released from that MTOC & carried to axon by motor proteins A. Overview B. Experimental Methods C. Microtubules 1. Structure 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs C. 5. Microtubules : MTOCS 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments Basal bodies & other MTOCs – Centrosomes are not the only MTOCs in cells; basal bodies at base of cilia & flagella serve as origin of ciliary & flagellar MTs; MTs grow out of them – Basal body cross-section looks like centriole; in fact, the two can give rise to one another – Sperm flagellum arises from basal body derived from sperm centriole that had been part of meiotic spindle of spermatocyte from which the sperm arose – Conversely, sperm basal body typically becomes centriole during fertilized egg's first mitotic division of fertilized egg A. Overview B. Experimental Methods C. Microtubules 1. Structure 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs C. 5. Microtubules : MTOCS 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments Plant MTOC - lack both centrioles & centrosomes; MTOCs more dispersed than those of animals – In plant endosperm cells, the primary MTOC consists of patches of material situated at outer surface of nuclear envelope from which cytoskeletal MTs emerge – MT nucleation also thought to occur throughout plant cell cortex A. Overview B. Experimental Methods C. Microtubules 1. Structure 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs C. 5. Microtubules : MTOCS 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments MT nucleation – Despite diverse appearances, all MTOCs play similar roles in all cells – Control number of MTs that form & their polarity – Control the number of protofilaments that make up their walls – Control the time & location of MT assembly A. Overview B. Experimental Methods C. Microtubules 1. Structure 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs C. 5. Microtubules : MTOCS 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments – – – – All MTOCs share a common protein component, gtubulin (discovered in mid-1980s); it is ~0.005% of total cell protein while a- & b-tubulins are 2.5% of total nonneural cell protein Fluorescent anti-g-tubulin antibodies (ABs) stain all MTOCs, like centrosome PCM; suggests it is critical component in MT assembly & nucleation Microinject anti-g-tubulin AB into living cell —> blocks MT reassembly after depolymerization by drugs or cold temperatures Genetically engineered fungi lacking functional gtubulin gene cannot assemble normal MTs A. Overview B. Experimental Methods C. Microtubules 1. Structure 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs C. 5. Microtubules : MTOCS 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments Nucleation mechanism revealed by structure/composition studies of PCM at centrosome periphery – Insoluble fibers of PCM are thought to serve as attachment sites for ring-shaped structures that have same diameter as MTs (25 nm) & contain g-tubulin – Ring-shaped structures found when centrosomes were purified & incubated with gold-labeled anti-g-tubulin ABs —> cluster in rings/semi-circles at MT minus ends (ends embedded in PCM) – Isolate similar ring-shaped complexes (g-TuRCs) from cell extracts; nucleate MT assembly in vitro A. Overview B. Experimental Methods C. Microtubules 1. Structure 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs C. 5. Microtubules : MTOCS 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments Model - helical array of 13 g-tubulin subunits forms open, ring-shaped template on which first row of ab-tubulin dimers assemble; – Only a-tubulin of heterodimer can bind to ring of gsubunits, establishing polarity of entire MT 2 other tubulin isoforms d-tubulin & e-tubulin have also been identified in centrosomes, but their function has not been determined A. Overview B. Experimental Methods C. Microtubules 1. Structure C. 6. Microtubules : Dynamic 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments 6. Dynamic properties of microtubules – – – – Alberts: Table 16-2; Fig 16-16, 16-17 MTs vary markedly in stability even though similar morphologically - spindle/cytoskeleton labile; mature neuron MTs less labile; cilia/flagella very stable; lability allows cell to respond to stimuli Cilia/flagella MTs are stabilized by MAP attachment & by enzymatic modification (e. g. acetylation) of specific amino acid residues within tubulin subunits Labile MTs in living cells can be disassembled without disrupting other cell structures via a number of treatments A. Overview B. Experimental Methods C. Microtubules 1. Structure C. 6. Microtubules : Dynamic 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments Treatments that cause MT disassembly; usually interfere with noncovalent bonds holding them together – – – – Cold temperatures Hydrostatic pressure Elevated Ca2+ concentration Variety of chemicals (often used in chemotherapy) - CO, vinblastine, vincristine, NO, podophyllotoxin A. Overview B. Experimental Methods C. Microtubules 1. Structure C. 6. Microtubules : Dynamic 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments Some treatments (taxol) disrupt MT dynamic activity & act by doing the opposite; inhibit disassembly – Taxol binds MT polymer & thus prevents disassembly; cell cannot build new MT structures A. Overview B. Experimental Methods C. Microtubules 1. Structure C. 6. Microtubules : Dynamic 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments Cytoskeletal MT lability reflects fact that they are polymers formed by noncovalent association of dimers; subject to depolymerization/repolymerization as cell needs change Dramatic changes in MT spatial organization may be achieved by combination of 2 separate mechanisms – Rearrangement of existing MTs – Disassembly of existing MTs & reassembly of new ones in different cell regions A. Overview B. Experimental Methods C. Microtubules 1. Structure C. 6. Microtubules : Dynamic 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments Study of MT dynamics in vitro - suggest that cytoskeleton can rapidly remodel & respond to stimuli – Early studies established that GTP binding to b -subunit required for MT assembly; GTP hydrolysis not needed for binding, but it is hydrolyzed soon after dimer attached to MT end; GDP stays bound – After dimer is released from MT during disassembly & enters soluble pool, GDP is replaced by GTP, thus recharging dimer so that it can add to polymer again – A GTP molecule is also bound to a-tubulin subunit, but it is not exchangeable & it is not hydrolyzed after subunit incorporation A. Overview B. Experimental Methods C. Microtubules 1. Structure C. 6. Microtubules : Dynamic 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments Assembly is not energetically inexpensive since it includes GTP hydrolysis, but it does allow the cell to control assembly & disassembly independently – A dimer being added to MT has a bound GTP; dimer being released from MT has bound GDP – GDP- & GTP dimers have different conformations & participate in different reactions; the ends of growing & shrinking MTs have different structures A. Overview B. Experimental Methods C. Microtubules 1. Structure C. 6. Microtubules : Dynamic 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments The above facts lead to the following model: – When a MT is growing, the "+" end is present as an open sheet to which GTP-dimers are added – During rapid growth periods, tubulin dimers are added faster than GTP can be hydrolyzed – The resultant cap of GTP-dimers on MT at protofilament ends is thought to favor the addition of more subunits & hence MT growth – However, MTs with open ends thought to undergo spontaneous reaction leading to tube closure A. Overview B. Experimental Methods C. Microtubules 1. Structure 2. MAPs 3. Functions C. 6. Microtubules : Dynamic 4. Microtubule Motors 5. MTOCs 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments – tube closure is accompanied by hydrolysis of bound GTP, changing tubulin dimer conformation —> resultant mechanical strain destabilizes MTs – Strain is released as protofilaments curl out from tubule & catastrophically depolymerize – Disassembly can occur remarkably fast, especially in vivo, which allows very rapid MT cytoskeleton disassembly A. Overview B. Experimental Methods C. Microtubules 1. Structure C. 6. Microtubules : Dynamic 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments Study of MT dynamics in vivo: dynamic character of MT cytoskeleton inside cell is best revealed by microinjecting labeled tubulin into nondividing cultured cell – Inject labeled tubulin into nondividing cultured cell — > labeled subunits rapidly incorporated into preexisting cytoskeleton MTs, even in absence of any obvious morphological change – Watch cell with fluorescent-labeled MTs over time —> some MTs grow, others shrink; dynamic – Both growth & shrinkage in vivo occur predominantly at "+" end of polymer, the end located opposite the centrosome (or other MTOC) A. Overview B. Experimental Methods C. Microtubules 1. Structure 2. MAPs 3. Functions C. 6. Microtubules : Dynamic 4. Microtubule Motors 5. MTOCs 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments – Single MTs switch randomly & unpredictably between growing & shrinking (dynamic instability) – MTs shrink faster than they grow, so in a matter of minutes, MTs disappear & are replaced by new MTs that grow out from centrosome A. Overview B. Experimental Methods C. Microtubules 1. Structure 2. MAPs C. 7. Microtubules: Flagella 3. Functions 4. Microtubule Motors 5. MTOCs 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments 7. Cilia and flagella structure – Alberts: Fig 16-80, 81, 82, 83, 84 Entire ciliary or flagellar projection is covered by membrane continuous with cell membrane Cilium core (axoneme) contains an array of MTs that run longitudinally through entire organelle – – Usually 9 peripheral doublet MTs surrounding central pair of single MTs; known as 9 + 2 pattern or array; all MTs in array have same polarity ("+" ends at tip, "-" ends at base) Doublets - 1 complete (A tubule; 13 subunits) MT; 1 incomplete (B tubule) MT with 10 or 11 subunits A. Overview B. Experimental Methods C. Microtubules 1. Structure 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs C. 7. Microtubules: Flagella 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments – Not all eukaryotes have them; cilia & flagella generally absent among fungi, nematodes & insects – Where they do occur, they nearly always show same 9 + 2 array, a reminder that all living eukaryotes have evolved from a common ancestor – Despite high degree of conservation (e. g. 9 + 2 pattern) some evolutionary departures: » 9 + 1 array in flatworms » 9 + 0 array in Asian horseshoe crab, eel, mayfly; some lacking central elements are motile, some not A. Overview B. Experimental Methods C. Microtubules 1. Structure 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs C. 7. Microtubules: Flagella 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments Central MTs enclosed by projections forming central sheath; sheath connected to doublet A MTs by radial spokes; doublets connected by interdoublet bridge made of elastic protein nexin Pair of arms (inner & outer) project from A MT in clockwise direction (when viewed base to tip) Radial spokes typically in groups of three with major repeat of 96 nm Inner & outer dynein arms staggered along A MT length (outer arms spaced every 24 nm; inner arms arranged to match unequal spacing of radial spokes) A. Overview B. Experimental Methods C. Microtubules 1. Structure 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs C. 7. Microtubules: Flagella 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments Cilia/flagellae emerge from basal bodies - 9 peripheral fibers consisting of 3 MTs rather than 2 (A tube complete, B/C incomplete); similar in structure to centrioles – No central MTs as in centrioles; also similar to centrioles in other ways – A & B tubules elongate to form cilia/flagella doublet; if sheared off, regrow from basal body A. Overview B. Experimental Methods C. Microtubules 1. Structure 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs C. 7. Microtubules: Flagella 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments The mechanism of ciliary & flagellar locomotion: sliding filament model suggested mechanism of ciliary/flagellar movement was sliding of adjacent MT doublets relative to one another In model, dynein arms act as swinging crossbridges that generate forces needed for ciliary/flagellar movement; dynein arms projecting from one doublet walk along adjacent doublet wall —> sliding A. Overview B. Experimental Methods C. Microtubules 1. Structure 2. MAPs 3. Functions 4. Microtubule Motors 5. MTOCs C. 7. Microtubules: Flagella 6. Dynamic Properties 7. Flagella and Cilia D. Microfilaments Sequence of events in ciliary/flagellar sliding motion – Dynein arms anchored on a doublet's A MT attach to binding sites on B MT of adjacent doublet – Dynein molecules undergo conformational change; causes A MT doublet to move slightly toward basal end of attached B MT doublet – Dynein then releases B tubule of adjacent doublet – Dynein arms reattach to adjacent doublet's B MT closer to its base so another cycle can begin A. Overview B. Experimental Methods C. Microtubules D. Microfilaments 1. Structure 2. P/D 3. Myosins 4. Muscle Contraction D. Microfilaments 5. Nonmuscle Actin 1. 2. 3. 4. 5. Structure Polymerization/depolymerization Myosin Muscle Contraction Non-muscle motility A. Overview B. Experimental Methods C. Microtubules D. Microfilaments 1. Structure 2. P/D D. 1. Microfilaments: Structure 3. Myosins 4. Muscle Contraction 5. Nonmuscle Actin 1. Structure Alberts Fig 16-12 Microfilaments: – – – – ~8 nm diameter made of globular actin subunits (G-actin) found in most animal cells, also higher plants “Microfilament,” “actin filament,” “F-actin filaments” are synonyms but F-actin often used for those formed in vitro A. Overview B. Experimental Methods C. Microtubules D. Microfilaments 1. Structure 2. P/D D. 1. Microfilaments: Structure 3. Myosins 4. Muscle Contraction 5. Nonmuscle Actin In presence of ATP, G-actin polymerizes to form stiff filament made of 2 strands of F-actin wound around each other in a helical configuration – Each subunit has polarity & all subunits are pointed in same direction, so entire MF has polarity Depending on cell type & activity in which it is engaged, MFs can be organized into highly ordered arrays, loose ill-defined networks, or tightly anchored bundles A. Overview B. Experimental Methods C. Microtubules D. Microfilaments 1. Structure 2. P/D D. 1. Microfilaments: Structure 3. Myosins 4. Muscle Contraction 5. Nonmuscle Actin Actin identified more than 50 years ago as one of major contractile proteins of muscle cells – Since then found to be major protein in virtually every eukaryotic cell examined – Higher plants & animals possess number of actincoding genes whose products are specialized for different types of motile processes – Actin structure highly conserved evolutionarily (yeast actin & rabbit skeletal actin 88% identical); means that nearly all aminos are crucial to function; actin from diverse sources can copolymerize A. Overview B. Experimental Methods C. Microtubules D. Microfilaments 1. Structure 2. P/D D. 1. Microfilaments: Structure 3. Myosins 4. Muscle Contraction 5. Nonmuscle Actin Actin detected microscopically: – By electron microscopy, using proteolytically cleaved myosin head fragments (HMM or S1 fragments) » HMM & S1 bind actin all along MF —> see polarity in EM; one end of MF pointed, other end barbed » Orientation of arrowheads formed by S1-actin complex provides information as to direction in which MFs are likely to be moved by myosin motor protein – By fluorescence microscopy, with fluorescently labeled S1 or anti-actin ABs A. Overview B. Experimental Methods C. Microtubules D. Microfilaments 1. Structure D. 2. Microfilaments: P/D 2. P/D 3. Myosins 4. Muscle Contraction 5. Nonmuscle Actin 2. MF polymerization/depolymerization Alberts Table 16 – 2, Fig. 16 – 36, 37, 38 Before polymerization, actin monomer binds to adenosine nucleotide (usually ATP); like GTP in MTs – – Actin is an ATPase (like tubulin is GTPase); role of ATP in MF assembly is same as GTP in MT Some time after incorporation into growing actin filament, ATP hydrolyzed to ADP If filaments built at high rate, the end has actinATP cap (hinders disassembly, favors assembly) A. Overview B. Experimental Methods C. Microtubules D. Microfilaments 1. Structure 2. P/D 3. Myosins 4. Muscle Contraction D. 2. Microfilaments: P/D 5. Nonmuscle Actin Actin polymerization can be studied in vitro by labeling or viscosity studies – In vitro with high concentration of labeled G-actin, both ends labeled but ..... » One end incorporates monomers at 5 - 10 times higher rate than the other end » Decoration with S1 myosin fragment reveals that barbed ("+" end) of MF is fast-growing end, while the pointed ("-") end is the slow-growing tip – In lower concentrations of G-actin: » Actin-ATP subunits add to "+" end & actin-ADP subunits tend to leave from "-" » Can be demonstrated by pulse-chase “treadmilling” experiments » Don’t know if treadmilling occurs in vivo A. Overview B. Experimental Methods C. Microtubules D. Microfilaments 1. Structure 2. P/D 3. Myosins 4. Muscle Contraction D. 2. Microfilaments: P/D 5. Nonmuscle Actin MFs maintain a dynamic equilibrium between monomeric & polymeric actin - can be influenced by a variety of different proteins Changes in local conditions in particular part of cell can push equilibrium either toward assembly or disassembly – allows cell to reorganize its MFs cytoskeleton by controlling this equilibrium – need such reorganization for dynamic processes (cell locomotion & shape changes, cytokinesis) A. Overview B. Experimental Methods C. Microtubules D. Microfilaments 1. Structure 2. P/D 3. Myosins 4. Muscle Contraction D. 2. Microfilaments: P/D 5. Nonmuscle Actin Actin-binding proteins in the cell affect the nucleation and polymerization rate of microfilaments – Formin: A dimeric protein that initiates nucleation by capturing two monomers of actin, then remains associated with the plus end of a rapidly growing microfolament – Thymosin: A protein that binds to actin monomers and inhibits nucleotide exchange or polymerization, keeping much of the available actin in an unpolymerized state – Profilin: A protien that competes with thymosin for binding to actin monomers; it binds opposite the ATP binding site on actin and promotes polymerization at the plus end of a growing filament A. Overview B. Experimental Methods C. Microtubules D. Microfilaments 1. Structure 2. P/D 3. Myosins 4. Muscle Contraction D. 2. Microfilaments: P/D 5. Nonmuscle Actin Inhibitors of microfilament polymerization/depolymerization used to study microfilament polymerization (Table 16 - 2 A. Overview B. Experimental Methods C. Microtubules D. Microfilaments 1. Structure 2. P/D D. 3. Microfilaments: Myosins 3. Myosins 4. Muscle Contraction 5. Nonmuscle Actin Myosins Alberts fig 16 – 54, 55, 56, 57, 60, 61, 65, 68, 69, 72 Myosin's sole known function is as motor for actin; 3. – – – Almost all motors known to interact with actin are members of myosin superfamily all of them move toward MF plus end (except for myosin VI) First isolated from mammalian skeletal muscle & then from wide variety of eukaryotic cells: protists, higher plants, nonmuscle animal cells, vertebrate cardiac & smooth muscle tissues A. Overview B. Experimental Methods C. Microtubules D. Microfilaments 1. Structure 2. P/D D. 3. Microfilaments: Myosins 3. Myosins 4. Muscle Contraction 5. Nonmuscle Actin Structure of myosins – All share characteristic motor (head) domain, which has a site that binds actin filament & one that binds & hydrolyzes ATP to drive the myosin motor – While head domains of myosins are similar, tail domains are highly divergent – Myosins also contain variety of low molecular weight (light) chains – Based on these construction differences divided into 2 large groups - conventional & unconventional A. Overview B. Experimental Methods C. Microtubules D. Microfilaments 1. Structure 2. P/D D. 3. Microfilaments: Myosins 3. Myosins 4. Muscle Contraction 5. Nonmuscle Actin Conventional (type II): – found in various muscle tissues, and also in a variety of nonmuscle cells (generate tension at focal contacts, cytokinesis) – Structure of myosin II molecules: 6 polypeptide chains (one pair of heavy chains, 2 pairs of light chains); organized in a way that produces a highly asymmetric protein with 3 sections » A pair of globular heads that contain the molecule’s catalytic site » A pair of necks, each consisting of a single, uninterrupted ahelix & 2 associated light chains » A single, long, rod-shaped tail formed by the intertwining of long a-helical sections of the 2 heavy chains to form an ahelical coiled-coil A. Overview B. Experimental Methods C. Microtubules D. Microfilaments 1. Structure 2. P/D D. 3. Microfilaments: Myosins 3. Myosins 4. Muscle Contraction 5. Nonmuscle Actin Immobilize isolated myosin heads (S1 fragments) on glass cover slip —> cause actin filament sliding – Single head domain has all of the machinery needed for motor activity The fibrous tail plays a structural role, allowing the protein to form filaments Light chain phosphorylation regulates assembly of myosin II into thick filaments Tail ends of myosin molecule point toward filament center; heads point toward ends (bipolar) – Polarity of filament reverses at its center A. Overview B. Experimental Methods C. Microtubules D. Microfilaments 1. Structure 2. P/D D. 3. Microfilaments: Myosins 3. Myosins 4. Muscle Contraction 5. Nonmuscle Actin Skeletal muscle myosin II filaments are highly stable smaller myosin II filaments (most nonmuscle cells) often display transient construction (assembling when & where needed, then disassembling) A. Overview B. Experimental Methods C. Microtubules D. Microfilaments 1. Structure 2. P/D D. 3. Microfilaments: Myosins 3. Myosins 4. Muscle Contraction 5. Nonmuscle Actin Unconventional myosins subdivided into at least 14 different types – Each type is presumed to have its own specialized functions – Several types may be present together in same cell Some functions of unconventional myosins – Amoeboid movement & phagocytosis (myosin I) – Movement of cytoplasmic vesicles & organelles (myosins I, V, & VI) – Stereocilia in cochlea hair cells of inner ear (myosin VIIa) A. Overview B. Experimental Methods C. Microtubules D. Microfilaments 1. Structure 2. P/D D. 4. Microfilaments: Muscle 3. Myosins 4. Muscle Contraction 5. Nonmuscle Actin 4. Muscle contraction Alberts 16 – 73, 74, 75, 76, 61, 77, 78 Skeletal muscle cell structure - highly unorthodox; cylindrical; 10 - 100 µm thick; up to 400 mm long – – Skeletal muscle cells are multinucleate (100s), the result of embryonic fusion of mononucleate myoblasts (premuscle cells); even myoblasts from distantly related animals fuse in culture Because of their properties, these cells are more appropriately called muscle fibers A. Overview B. Experimental Methods C. Microtubules D. Microfilaments D. 4. Microfilaments: Muscle 1. Structure 2. P/D 3. Myosins 4. Muscle Contraction 5. Nonmuscle Actin Muscle fibers may have the most orderly internal structure of any cell in body – Muscle fiber is cable made up of hundreds of thinner, cylindrical strands (myofibrils) – Each myofibril is repeating linear array of contractile units (sarcomeres) – Each sarcomere, in turn, has characteristic banding pattern that gives muscle fiber a striated look Myofibrils separated by cytoplasm with intracellular membranes & mitochondria, lipid droplets, glycogen granules A. Overview B. Experimental Methods C. Microtubules D. Microfilaments D. 4. Microfilaments: Muscle 1. Structure 2. P/D 3. Myosins 4. Muscle Contraction 5. Nonmuscle Actin Banding pattern is result of partial overlap between thick & thin filaments – Each sarcomere extends from Z line to Z line (~2.5 µm) & contains several dark bands & light zones; there is a pair of light staining I bands at each end of sarcomere – More densely staining A band is between outer I bands; lightly staining H zone in A band center – Densely staining M line lies in center of H zone – I bands - only thin filaments; H zone - only thick filaments; A band outside H zone - both overlap A. Overview B. Experimental Methods C. Microtubules D. Microfilaments D. 4. Microfilaments: Muscle 1. Structure 2. P/D 3. Myosins 4. Muscle Contraction 5. Nonmuscle Actin Composition & organization of thin filaments – Thin filaments mostly actin – In addition to actin, thin filaments also contain two other proteins: troponin & tropomyosin » Tropomyosin - elongated, ~40 nm long; fits securely into grooves between two thin filament actin chains; each rodshaped tropomyosin interacts with 7 actin subunits linearly along F-actin chain » Troponin - globular protein complex with 3 subunits - each has distinct, important functional role; ~40 nm apart on thin filament, contact both actin & tropomyosin thin filament components – Actin filaments of each half sarcomere aligned with barbed ends linked to Z line by a-actinin A. Overview B. Experimental Methods C. Microtubules D. Microfilaments D. 4. Microfilaments: Muscle 1. Structure 2. P/D 3. Myosins 4. Muscle Contraction 5. Nonmuscle Actin Composition & organization of thick filaments – Thick filaments are several 100 myosins & a small number of other proteins – Like filaments formed in vitro, thick filament polarity is reversed at sarcomere center; filament center is composed of opposing tail regions of myosin molecules & is devoid of heads – Myosin heads project from each thick filament along remainder of its length due to staggered positions of myosins making up the body of the filament A. Overview B. Experimental Methods C. Microtubules D. Microfilaments 1. Structure 2. P/D D. 4. Microfilaments: Muscle 3. Myosins 4. Muscle Contraction 5. Nonmuscle Actin – Titin (~3 x 106 dalton MW [27,000 aa’s], 1 µm long) - largest protein yet discovered; originates at M line & extends along myosin filament past A band to terminate on Z line » Highly elastic protein; stretches (bungee cord) as certain domains within molecule are unfolded » It is thought to prevent sarcomere from being pulled apart during muscle stretching » Also maintains myosin filaments in proper position in sarcomere center during contraction A. Overview B. Experimental Methods C. Microtubules D. Microfilaments 1. Structure 2. P/D 3. Myosins 4. Muscle Contraction 5. Nonmuscle Actin D. 4. Microfilaments: Muscle The Sliding Filament Model of muscle contraction – muscles contract by shortening at sarcomere level – combined decrease in sarcomere length accounts for decrease in length of entire muscle – Sarcomere banding patterns shift during contraction shows that filaments slide » H zone & I bands decrease in width during contraction & eventually disappear; A bands stay same » During contraction, Z lines move inward & approach A band outer edges until they touch each other A. Overview B. Experimental Methods C. Microtubules D. Microfilaments D. 4. Microfilaments: Muscle 1. Structure 2. P/D 3. Myosins 4. Muscle Contraction 5. Nonmuscle Actin Molecular basis of contraction – During contraction, each myosin head extends out & binds tightly to thin filaments forming cross-bridges – Once bridges form, heads change shape, bending toward sarcomere center (power stroke) – Moves actin filament over thick filament (~5 - 15 nm; controversy exists over these distances) toward sarcomere center – Heads hydrolyze ATP & act as levers for thin filament motion; each myosin cross-bridge mechanical activity cycle is accompanied by ATPase activity cycle A. Overview B. Experimental Methods C. Microtubules D. Microfilaments D. 4. Microfilaments: Muscle 1. Structure 2. P/D 3. Myosins 4. Muscle Contraction 5. Nonmuscle Actin Details of the sliding filament model – Cycle starts with ATP binding to myosin head, which induces dissociation of cross-bridge from actin – ATP binding followed by its hydrolysis before head contacts actin filament; ADP & Pi products stay bound at enzyme active site – Energy released by hydrolysis absorbed by myosin as a whole, placing the cross-bridge in energized state, like a stretched spring capable of spontaneous movement – Energized myosin attaches to actin & releases its bound Pi A. Overview B. Experimental Methods C. Microtubules D. Microfilaments D. 4. Microfilaments: Muscle 1. Structure 2. P/D 3. Myosins 4. Muscle Contraction 5. Nonmuscle Actin – Triggers a large conformational change, driven by stored free energy – The shape change shifts actin filament toward sarcomere center (myosin head power stroke) – Bound ADP leaves & new ATP attaches, inducing release of cross-bridge; cycle starts over – In absence of ATP, cross-bridges stay tightly intact binding actin (causes rigor mortis after death) Skeletal muscle contraction is triggered by a nerve-impulse mediated release of calcium ions into the sarcomere A. Overview B. Experimental Methods C. Microtubules D. Microfilaments 1. Structure 2. P/D D. 4. Microfilaments: Muscle 3. Myosins 4. Muscle Contraction 5. Nonmuscle Actin Regulation of skeletal muscle contraction – Muscle fibers are organized into motor units; all fibers of motor unit jointly innervated by branches of one motor neuron; contract simultaneously if stimulated by impulse transmitted along that neuron » Point where neuron axon terminus & muscle fiber make contact called neuromuscular junction » Junction is site of nerve impulse transmission from axon across synaptic cleft to muscle fiber » Muscle fiber plasma membrane is also excitable & capable of conducting an action potential A. Overview B. Experimental Methods C. Microtubules D. Microfilaments 1. Structure 2. P/D D. 4. Microfilaments: Muscle 3. Myosins 4. Muscle Contraction 5. Nonmuscle Actin – Unlike a neuron, where an action potential stays at the cell surface, skeletal muscle cell impulse is propagated into cell interior along membranous folds (transverse [T] tubules) » T tubules terminate in close proximity to a cytoplasmic membrane system (sarcoplasmic reticulum [SR]) that forms membranous sleeve around myofibril (specialized form of smooth ER) » ~80% of SR membrane integral protein is ATPdriven Ca2+ pump (moves Ca2+ from cytosol into SR lumen where it is stored until it is needed) A. Overview B. Experimental Methods C. Microtubules D. Microfilaments 1. Structure 2. P/D 3. Myosins D. 4. Microfilaments: Muscle 4. Muscle Contraction 5. Nonmuscle Actin – With arrival of action potential at SR via T tubules, opens Ca2+ channels in SR membrane are opened » Ca2+ diffuses out of SR compartment & over short distance to myofibrils » [Ca+2] levels go from ~2 x 10-7 M to ~5 x 10-5 M – In a relaxed sarcomere (low [Ca2+]): » Tropomyosin blocks myosin-binding sites on actin molecules » Tropomyosin’s position in filament groove blocking sites is controlled by troponin A. Overview B. Experimental Methods C. Microtubules D. Microfilaments 1. Structure 2. P/D D. 4. Microfilaments: Muscle 3. Myosins 4. Muscle Contraction 5. Nonmuscle Actin – When [Ca2+] rises: » Ca2+ binds to troponin C subunits » This causes a conformational change in troponin » And this causes adjacent tropomyosin to move ~1.5 nm closer to center of filament’s groove – The movement of tropomyosin exposes myosin-binding site on adjacent actins, crossbridges form, contraction occurs – Each troponin controls position of 1 tropomyosin, which, in turn, controls binding capacity of 7 adjacent actin monomers A. Overview B. Experimental Methods C. Microtubules D. Microfilaments 1. Structure 2. P/D D. 4. Microfilaments: Muscle 3. Myosins 4. Muscle Contraction 5. Nonmuscle Actin – When stimulation from motor neuron stops: » SR Ca2+ channels close & excess Ca2+ pumped from cytosol back into SR by the Ca2+ pump » As [Ca2+] decreases, Ca2+ ions dissociate from troponin binding sites » Tropomyosin molecules return to their original positions and block actin-myosin binding A. Overview B. Experimental Methods C. Microtubules D. Microfilaments 1. Structure 2. P/D D. 5. Microfilaments: Nonmuscle 3. Myosins 4. Muscle Contraction 5. Nonmuscle Actin Some examples of nonmuscle actin-myosin 5. – – – – – – – – – – Alberts Fig. 16 – 50, 51, 52 Cytokinesis Phagocytosis Cytoplasmic streaming (directed bulk flow of cytoplasm occurring in certain large plant cells) Vesicle trafficking Blood platelet activation Lateral movements of integral proteins within membranes Cell substratum interactions, cell locomotion & axonal outgrowth Changes in cell shape Cell projections such as microvilli & stereocilia