AP Biology Discussion Notes
advertisement

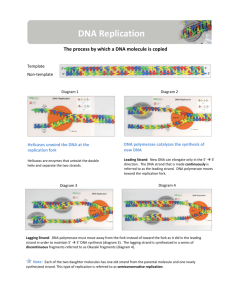
AP Biology Discussion Notes Wednesday 1/27/2016 Goals for Today • Be able to say what the central dogma of biology is. • Be able to describe the structure of DNA and its associated terms • Be able to describe the process & players in DNA replication • Be able to “build” a strand of DNA Question of the Day 1/27 • What is DNA replication & what can be said about this process & what does that mean? (Is it liberal?) Question of the Day 1/27 • What is DNA replication & what can be said about this process & what does that mean? (Is it liberal?) Figure 16.9-1 Semiconservative Replication A T C G T A A T G C (a) Parent molecule Figure 16.9-2 Semiconservative Replication A T A T C G C G T A T A A T A T G C G C (a) Parent molecule (b) Separation of strands Figure 16.9-3 Semiconservative Replication A T A T A T A T C G C G C G C G T A T A T A T A A T A T A T A T G C G C G C G C (a) Parent molecule (b) Separation of strands (c) “Daughter” DNA molecules, each consisting of one parental strand and one new strand Chapter 16 in your book Figure 16.10 (a) Conservative model (b) Semiconservative model (c) Dispersive model Parent cell First replication Second replication • Experiments by Matthew Meselson and Franklin Stahl supported the semiconservative model • They labeled the nucleotides of the old strands with a heavy isotope of nitrogen 15N, while any new nucleotides were labeled with a lighter isotope, 14N Figure 16.11a EXPERIMENT 2 Bacteria transferred to medium with 14N (lighter isotope) 1 Bacteria cultured in medium with 15N (heavy isotope) RESULTS 3 DNA sample centrifuged after first replication ? 4 DNA sample centrifuged after second ? replication Less dense More dense Figure 16.11b CONCLUSION Predictions: First replication Conservative model Semiconservative model Dispersive model Second replication DNA Replication: A Closer Look • The copying of DNA is remarkable in its speed and accuracy –E.coli has about 4.6 Million nucleotide/base pairs and replicates DNA, then divides into 2 new cells in less than an hour! –Humans have ~6 Billion nucleotide/base pairs and replicate their DNA in a few hours Getting Started • Replication begins at particular sites called origins of replication, where the two DNA strands are separated, opening up a replication “bubble” • A eukaryotic chromosome may have hundreds or even thousands of origins of replication • Replication proceeds in both directions from each origin, until the entire* molecule is copied Figure 16.12a (a) Origin of replication in an E. coli cell Origin of replication Parental (template) strand Daughter (new) strand Doublestranded DNA molecule Replication bubble Replication fork Two daughter DNA molecules 0.5 m Synthesizing a New DNA Strand • Enzymes called DNA polymerases catalyze the elongation of new DNA at a replication fork • The rate of elongation is about 500 nucleotides per second in bacteria and 50 per second in human cells • How do we speed this up in our cells? Figure 16.12b (b) Origins of replication in a eukaryotic cell Double-stranded Origin of replication DNA molecule Parental (template) strand Bubble Daughter (new) strand Replication fork Two daughter DNA molecules 0.25 m Can you answer these questions: • In which direction is each strand being built? • How is it built? • Where is it moving towards? • What enzymes are involved and what are their roles? Figure 16.15a Leading strand Overview Origin of replication Lagging strand Primer Lagging strand Overall directions of replication Leading strand DNA Replication • At the end of each replication bubble is a replication fork, a Y-shaped region where new DNA strands are elongating • Helicases are enzymes that untwist the double helix at the replication forks • Single-strand binding proteins bind to and stabilize single-stranded DNA • Topoisomerase corrects “over winding” ahead of replication forks by breaking, swiveling, and rejoining DNA strands Figure 16.15a Leading strand Overview Origin of replication Lagging strand Primer Lagging strand Overall directions of replication Leading strand What enzyme? • What enzyme will build the new DNA strand (the new polymer of DNA)? There is a small problem… • Besides ONLY being able to build in the 5’ to 3’ direction, DNA polymerases cannot initiate synthesis of a polynucleotide; they can only add nucleotides to the 3 end The Solution…. • The initial nucleotide strand is a short RNA primer Figure 16.15a Leading strand Overview Origin of replication Lagging strand Primer Lagging strand Overall directions of replication Leading strand Antiparallel Elongation • The antiparallel structure of the double helix affects replication • DNA polymerases add nucleotides only to the free 3end of a growing strand; therefore, a new DNA strand can elongate only in the 5to 3direction • This creates a “leading” and a “lagging” strand Let’s try to understand this…. Can you answer these questions: • In which direction is each strand being built? • How is it built? • Where is it moving towards? • What enzymes are involved & what are their roles? Figure 16.15a Leading strand Overview Origin of replication Lagging strand Primer Lagging strand Overall directions of replication Leading strand Antiparallel Elongation • Along one template strand of DNA, the DNA polymerase synthesizes a leading strand continuously, moving toward the replication fork Figure 16.15 Leading strand Overview Origin of replication Lagging strand Primer Lagging strand Leading strand Overall directions of replication Origin of replication 3 5 RNA primer 5 3 3 Sliding clamp DNA pol III Parental DNA 5 3 5 5 3 3 5 Figure 16.15b Origin of replication 3 5 RNA primer 5 3 3 Sliding clamp DNA pol III Parental DNA 5 3 5 5 3 3 5 Antiparallel Elongation • To elongate the other new strand, called the lagging strand, DNA polymerase must work in the direction away from the replication fork • The lagging strand is synthesized as a series of segments called Okazaki fragments, which are joined together by DNA ligase Figure 16.16 3 Overview 5 Template strand 3 Leading strand 3 Origin of replication 5 RNA primer for fragment 1 Lagging strand 2 5 1 3 5 3 5 Okazaki fragment 1 RNA primer for fragment 2 5 Okazaki 3 fragment 2 2 Lagging strand 1 3 5 1 5 3 5 3 2 1 3 5 5 3 2 1 3 5 Overall direction of replication 1 Overall directions of replication Leading strand Figure 16.16a Overview Leading strand Origin of replication Lagging strand Lagging strand 2 1 Overall directions of replication Leading strand Figure 16.17 Overview Origin of replication Leading strand Leading strand Lagging strand Overall directions of replication Lagging strand Leading strand DNA pol III 5 3 3 Parental DNA Primer 5 3 Primase 5 DNA pol III 4 Lagging strand DNA pol I 35 3 2 DNA ligase 1 3 5 Figure 16.17a Overview Leading strand Lagging strand Origin of replication Lagging strand Leading strand Overall directions of replication Leading strand DNA pol III 5 3 3 Parental DNA Primer 5 3 Primase Figure 16.17b Overview Origin of replication Leading strand Leading strand Lagging strand Overall directions of replication Lagging strand Leading strand Primer 5 DNA pol III 4 3 Lagging strand DNA pol I 35 3 2 DNA ligase 1 3 5 Figure 16.18 The DNA Replication Complex DNA pol III Parental DNA 5 3 5 3 3 5 5 Connecting protein 3 Helicase 3 DNA pol III 5 Leading strand 3 5 Lagging strand Lagging strand template Proofreading and Repairing DNA • DNA polymerases proofread newly made DNA, replacing any incorrect nucleotides Figure 16.19 5 3 3 5 Nuclease 5 3 3 5 DNA polymerase 5 3 3 5 DNA ligase 5 3 3 5 Evolutionary Significance of Altered DNA Nucleotides • Error rate after proofreading repair is low but not zero • Sequence changes may become permanent and can be passed on to the next generation (when would they be passed on?) • These changes (mutations) are the source of the genetic variation upon which natural selection operates “The Players” in DNA Replication • The Enzymes – Helicase (“Hacks”) – DNA Polymerase (“Pastes”) – Ligase (“Links”) • • • • • • Nucleotides (A,T,C,G) Origin(s) of Replication Leading Strand Lagging Strand 5’ 3’ Okazaki Fragments Summary of DNA Replication Helicase unwinds and unzips DNA at O.O.R. LOTS of OOR’s/Chrom. in Euk’s: ONE in Prok’s. DNA Polymerase jumps in at O.O.R. and adds new nucleotides from 5’3’ LEADING and LAGGING strands formed from each template strand LAGGING strand Okazaki Fragments are connected by Ligase Now have two copies of the same DNA! Can you answer these questions: • In which direction is each strand being built? • How is it built? • Where is it moving towards? • What enzymes are involved? DNA assignment