Chapter 14
advertisement

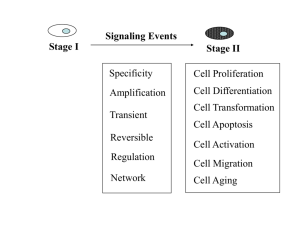
Chapter 14 Principles of Cell Signaling By Melanie H. Cobb & Elliott M. Ross 14.2 Cellular signaling is primarily chemical • Cells can detect both chemical and physical signals. • Physical signals are generally converted to chemical signals at the level of the receptor. 14.3 Receptors sense diverse stimuli but initiate a limited repertoire of cellular signals • Receptors contain: – a ligand-binding domain – an effector domain • Receptor modularity allows a wide variety of signals to use a limited number of regulatory mechanisms. 14.3 Receptors sense diverse stimuli but initiate a limited repertoire of cellular signals • Cells may express different receptors for the same ligand. • The same ligand may have different effects on the cell depending on the effector domain of its receptor. 14.4 Receptors are catalysts and amplifiers • Receptors act by increasing the rates of key regulatory reactions. • Receptors act as molecular amplifiers. 14.5 Ligand binding changes receptor conformation • Receptors can exist in active or inactive conformations. • Ligand binding drives the receptor toward the active conformation. 14.6 Signals are sorted and integrated in signaling pathways and networks • Signaling pathways usually have multiple steps and can diverge and/or converge. • Divergence allows multiple responses to a single signal. • Convergence allows signal integration and coordination. 14.7 Cellular signaling pathways can be thought of as biochemical logic circuits • Signaling networks are composed of groups of biochemical reactions. – The reactions function as mathematical logic functions to integrate information. • Combinations of such logic functions combine as signaling networks to process information at more complex levels. 14.8 Scaffolds increase signaling efficiency and enhance spatial organization of signaling • Scaffolds: – organize groups of signaling proteins – may create pathway specificity by sequestering components that have multiple partners 14.8 Scaffolds increase signaling efficiency and enhance spatial organization of signaling • Scaffolds increase the local concentration of signaling proteins. • Scaffolds localize signaling pathways to sites of action. 14.9 Independent, modular domains specify protein-protein interactions • Protein interactions may be mediated by small, conserved domains. • Modular interaction domains are essential for signal transmission. • Adaptors consist exclusively of binding domains or motifs. 14.10 Cellular signaling is remarkably adaptive • Sensitivity of signaling pathways is regulated to allow responses to change over a wide range of signal strengths. • Feedback mechanisms execute this function in all signaling pathways. 14.10 Cellular signaling is remarkably adaptive • Most pathways contain multiple adaptive feedback loops to cope with signals of various strengths and durations. 14.11 Signaling proteins are frequently expressed as multiple species • Distinct species (isoforms) of similar signaling proteins expand the regulatory mechanisms possible in signaling pathways. 14.11 Signaling proteins are frequently expressed as multiple species • Isoforms may differ in: – function – susceptibility to regulation – expression • Cells may express one or several isoforms to fulfill their signaling needs. 14.12 Activating and deactivating reactions are separate and independently controlled • Activating and deactivating reactions are usually executed by different regulatory proteins. • Separating activation and inactivation allows for fine-tuned regulation of amplitude and timing. 14.13 Cellular signaling uses both allostery and covalent modification • Allostery refers to the ability of a molecule to alter the conformation of a target protein when it binds noncovalently to that protein. • Modification of a protein’s chemical structure is also frequently used to regulate its activity. 14.14 Second messengers provide readily diffusible pathways for information transfer • Second messengers can propagate signals between proteins that are at a distance. • cAMP and Ca2+ are widely used second messengers. 14.15 Ca2+ signaling serves diverse purposes in all eukaryotic cells • Ca2+ serves as a second messenger and regulatory molecule in essentially all cells. 14.15 Ca2+ signaling serves diverse purposes in all eukaryotic cells • Ca2+ acts directly on many target proteins. – It also regulates the activity of a regulatory protein calmodulin. • The cytosolic concentration of Ca2+ is controlled by organellar sequestration and release. 14.16 Lipids and lipid-derived compounds are signaling molecules • Multiple lipid-derived second messengers are produced in membranes. • Phospholipase Cs release soluble and lipid second messengers in response to diverse inputs. 14.16 Lipids and lipid-derived compounds are signaling molecules • Channels and transporters are modulated by different lipids in addition to inputs from other sources. • PI 3-kinase synthesizes PIP3 to modulate cell shape and motility. • PLD and PLA2 create other lipid second messengers. 14.17 PI 3-kinase regulates both cell shape and the activation of essential growth and metabolic functions • Phosphorylation of some lipid second messengers changes their activity. • PIP3 is recognized by proteins with a pleckstrin homology domain. 14.18 Signaling through ion channel receptors is very fast • Ion channels allow the passage of ions through a pore. – This results in rapid (microsecond) changes in membrane potential. 14.18 Signaling through ion channel receptors is very fast • Channels are selective for particular ions or for cations or anions. • Channels regulate intracellular concentrations of regulatory ions, such as Ca2+. 14.19 Nuclear receptors regulate transcription • Nuclear receptors modulate transcription by binding to distinct short sequences in chromosomal DNA known as response elements. 14.19 Nuclear receptors regulate transcription • Receptor binding to other receptors, inhibitors, or coactivators leads to complex transcriptional control circuits. • Signaling through nuclear receptors is relatively slow, consistent with their roles in adaptive responses. 14.20 G protein signaling modules are widely used and highly adaptable • The basic module is: – a receptor – a G protein – an effector protein 14.20 G protein signaling modules are widely used and highly adaptable • Cells express several varieties of each class of proteins. • Effectors are heterogeneous and initiate diverse cellular functions. 14.21 Heterotrimeric G proteins regulate a wide variety of effectors • G proteins convey signals by regulating the activities of multiple intracellular signaling proteins known as effectors. • Effectors are structurally and functionally diverse. 14.21 Heterotrimeric G proteins regulate a wide variety of effectors • A common G-protein binding domain has not been identified among effector proteins. • Effector proteins integrate signals from multiple G protein pathways. 14.22 Heterotrimeric G proteins are controlled by a regulatory GTPase cycle • Heterotrimeric G proteins are activated when the Gα subunit binds GTP. • GTP hydrolysis to GDP inactivates the G protein. 14.22 Heterotrimeric G proteins are controlled by a regulatory GTPase cycle • GTP hydrolysis is slow, but is accelerated by proteins called GAPs. • Receptors promote activation by allowing GDP dissociation and GTP association. – Spontaneous exchange is very slow. • RGS proteins and phospholipase C-βs are GAPs for G proteins. 14.23 Small, monomeric GTPbinding proteins are multiuse switches • Small GTP-binding proteins are: – active when bound to GTP – inactive when bound to GDP • GDP/GTP exchange catalysts known as GEFs (guanine nucleotide exchange factors) promote activation. 14.23 Small, monomeric GTPbinding proteins are multiuse switches • GAPs accelerate hydrolysis and deactivation. • GDP dissociation inhibitors (GDIs) slow spontaneous nucleotide exchange. 14.24 Protein phosphorylation/ dephosphorylation is a major regulatory mechanism in the cell • Protein kinases are a large protein family. • Protein kinases phosphorylate: – Ser and Thr – or Tyr – or all three 14.24 Protein phosphorylation/ dephosphorylation is a major regulatory mechanism in the cell • Protein kinases may recognize the primary sequence surrounding the phosphorylation site. • Protein kinases may preferentially recognize phosphorylation sites within folded domains. 14.25 Two-component protein phosphorylation systems are signaling relays • Two-component signaling systems are composed of sensor and response regulator components. 14.25 Two-component protein phosphorylation systems are signaling relays • Upon receiving a stimulus, sensor components undergo autophosphorylation on a histidine (His) residue. • Transfer of the phosphate to an aspartyl residue on the response regulator serves to activate the regulator. 14.26 Pharmacological inhibitors of protein kinases may be used to understand and treat disease • Protein kinase inhibitors are useful both: – for signaling research – as drugs • Protein kinase inhibitors usually bind in the ATP binding site. 14.27 Phosphoprotein phosphatases reverse the actions of kinases and are independently regulated • Phosphoprotein phosphatases reverse the actions of protein kinases. 14.27 Phosphoprotein phosphatases reverse the actions of kinases and are independently regulated • Phosphoprotein phosphatases may dephosphorylate: – phosphoserine/threonine – phosphotyrosine – or all three • Phosphoprotein phosphatase specificity is often achieved through the formation of specific protein complexes. 14.18 Covalent modification by ubiquitin and ubiquitinlike proteins is another way of regulating protein function • Ubiquitin and related small proteins may be covalently attached to other proteins as a targeting signal. • Ubiquitin is recognized by diverse ubiquitin binding proteins. 14.18 Covalent modification by ubiquitin and ubiquitinlike proteins is another way of regulating protein function • Ubiquitination can cooperate with other covalent modifications. • Ubiquitination regulates signaling in addition to its role in protein degradation. 14.29 The Wnt pathway regulates cell fate during development and other processes in the adult • Seven transmembrane-spanning receptors may control complex differentiation programs. • Wnts are lipid-modified ligands. 14.29 The Wnt pathway regulates cell fate during development and other processes in the adult • Wnts signal through multiple distinct receptors. • Wnts suppress degradation of βcatenin, a multifunctional transcription factor. 14.30 Diverse signaling mechanisms are regulated by protein tyrosine kinases • Many receptor protein tyrosine kinases are activated by growth factors. • Mutations in receptor tyrosine kinases can be oncogenic. 14.30 Diverse signaling mechanisms are regulated by protein tyrosine kinases • Ligand binding promotes: – receptor oligomerization – autophosphorylation • Signaling proteins bind to the phosphotyrosine residues of the activated receptor. 14.31 Src family protein kinases cooperate with receptor protein tyrosine kinases • Src is activated by release of intrasteric inhibition. • Activation of Src involves liberation of modular binding domains for activationdependent interactions. • Src often associates with receptors, including receptor tyrosine kinases. 14.32 MAPKs are central to many signaling pathways • MAPKs are activated by Tyr and Thr phosphorylation. • The requirement for two phosphorylations creates a signaling threshold. • The ERK1/2 MAPK pathway is usually regulated through Ras. 14.33 Cyclin-dependent protein kinases control the cell cycle • The cell cycle is regulated by cyclindependent protein kinases (CDKs). • Activation of CDKs involves: – protein binding – dephosphorylation – phosphorylation 14.34 Diverse receptors recruit protein tyrosine kinases to the plasma membrane • Receptors that bind protein tyrosine kinases use combinations of effectors similar to those used by receptor tyrosine kinases. • These receptors often bind directly to transcription factors.