401Lecture7Sp2013post
advertisement

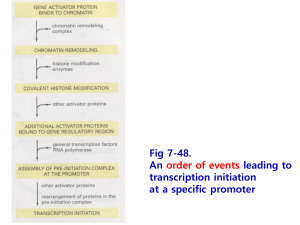
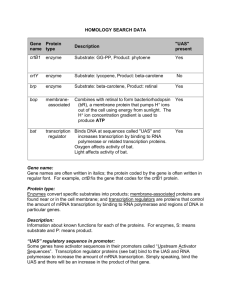
Outline • Questions from last lecture? • P. 40 questions on Pax6 gene • Mechanism of Transcription Activation – Transcription Regulatory elements • Comparison between yeast and mammals • Examples of mammalian transcriptional control regions – Transcription Factors (activators and repressors) • Mechanisms of Transcription repression – Comparison between prok and euk repression – Novel mechanism of repression by non-coding RNAs Different Pax 6 enhancers are active in different tissues: Results in different sites of transcription initiation and alternative splicing in different tissues Transcript a is detected in lense and pancreas. Lacks exons 1 and α Fig. 7-7 Lodish et al. 2008 Alternative Pax 6 promoters & enhancers are used to generate different transcripts in different tissues Transcript c: detected in retina lacks exons 0-4 contains exon α exon α is unique to vertebrates Fig. 7-7 Pax 6 Questions p. 40 1. What parts of the Pax6 gene contain regulatory elements? 2. What DNA constructs could have been used to generate the transgenic mice shown in the bottom panels of Fig. 7-7? 3. Could this same information have been obtained from an in vitro cell culture experiment? Why or why not? 4. What experiments could you perform to identify important transcription control regions? How would you know where to start looking? Linker-scanning Mutation of a Gene Regulatory Region Reporter gene activity What can you tell from comparing mutants 1, 2, 3 & 4? Fig. 7-21 Readings Fig 7.8 – example of how a SALLI gene enhancer was identified in an intergenic region 500 kb downstream of the SALL1 gene Techniques to Characterize CisActing Regulatory Regions • Transgenic mice – Create DNA constructs containing potential cis-acting sequences + reporter gene and introduce into embryonic stem cells • Reporter Gene Assay (fig. 7-25) – Cell culture approach • Linker Scanning Mutation Analysis (fig. 7-21) – In vivo or in cell culture • DNase I Footprinting (fig. 7-23) • Comparative genomics Purple = in text book readings Review at home: Techniques to Identify Regulatory Elements Comparison of Eukaryotic Control Regions Structure of a Transcription Factor Fig. 7-22. Lodish Conclusions • Different control regions can control transcription of the same gene in different cell types • Different subsets of transcription factors bind to control regions of the same gene in different cell types • Control regions can be located far from transcription start sites Transcription Initiation Requires Binding of Many Different Proteins to Enhancers and Promoters What are 3 different functions of proteins that regulate transcription by binding to specific DNA sequences? 1. 2. 3. Transcription activators Transcription repressors General transcription factors e.g. TATA binding protein At home: draw a picture of a eukaryotic promoter at the time of transcription initiation (include all proteins expected to be there) Example of a Typical Mammalian Enhancer Enhanceosome on the β-interferon enhancer What determines whether a given protein is found associated with the enhancer? • WT1 represses EGR-1 • SRF/TCF and AP1 are transcription activators • Based on this schematic, how is the mechanism of action different for the eukaryotic WT1 (Wilms Tumor) repressor compared to prokaryotic repressors (e.g. lac i)? Novel mechanisms for Gene Repression: Epigenetic Silencing of the Flowering Gene FLC by a Noncoding RNA Luciferase activity COLDAIR transcription is induced by cold COLDAIR noncoding RNA binds Polycomb group proteins FLC locus is methylated on histone H3 Silencing Science 331: 76-79 (2011) Summary • Activators promote transcription and are modular proteins composed of a DNA binding domain and an activation domain • Repressors inhibit transcription and are modular proteins composed of a DNA binding domain and a repressor domain • A cell must produce the specific set of activators required for transcription of a particular gene in order to express that gene • Noncoding RNAs can recruit proteins that promote silencing (e.g. histone methyl transferases)_