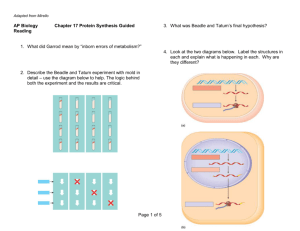
Transcription - ITG
advertisement

Basic transcription mechanisms II Thomas Dickmeis Institut für Toxikologie und Genetik, KIT, Karlsruhe thomas.dickmeis@kit.edu 1 Moodle and exam • Moodle: talks will be uploaded soon – Password this year: IntroGen13 • Exam: – Nick will set up a poll after Christmas to fix the date of the exam – Please send an email with your name and Matrikelnummer to nicholas.foulkes@kit.edu 2 Eukaryotic Transcription 3 Transcription in prokaryotes vs. eukaryotes 4 Stryer 2002 three important differences • Chromatin is the template (bacteria: „naked“ DNA) • Polymerase needs general transcription factors (GTFs) for promoter binding and initiation (bacteria: holoenzyme binds directly) • three polymerases (bacteria: one): – RNA pol I: 18S/28S rRNA – RNA pol II: mRNA, few small RNAs – RNA pol III: tRNA, 5S rRNA, other small RNAs 5 Transcription in prokaryotes vs. eukaryotes eubacteria Nucleus archaebacteria eukaryotes - - + Transcription and translation not separated not separated separated Genome organisation one circular chromosome one circular chromosome several linear chromosomes - - + few few majority operons + + - (exceptions?) Introns - - + RNA polymerases 1 1 3: Pol I = rRNA Pol II= mRNA Pol III = tRNA eubacterial archaebacterial archaebacterial 5´Cap on mRNA - - + polyA 3´ on mRNA - rare + Histones/ nucleosomes Non-coding sequences RNA polymerase type 6 Typical regulatory sequences of a Pol II transcribed gene Promoter: - binds GTFs Enhancer: - binds transcriptional regulators - increases promoter utilization - can be upstream, inside the gene or downstream („distal“ enhancers can be very far away) - orientation not important - often target for tissue-specific or temporal regulation Silencer: same, but decreases promoter utilization (How can distant sequences influence the promoter?) 7 DNA looping brings enhancers and promoters together (How can one prove this?) 8 Cooper 2000 The 3C technique allows the study of chromatin looping „3C“ = Chromosome Conformation Capture (How can one avoid intermolecular ligation?) (What has to be known in this case?) 9 Chromatin and transcription 10 Alberts 2002 Principles of enhancer function: I. making the promoter accessible ATP ADP+P 1. Chromatin remodelling makes the promoter accessible (How does one know if an octamer has been displaced?) 11 (finding the accessible chromatin: mapping of DNAseI hypersensitive sites) 12 Principles of enhancer function: I. making the promoter accessible 2. Chromatin modifications Generation of chromatin marks that can be bound e.g. by the basal transcription factors 13 Principles of enhancer function: II. Architectural proteins Bending of the DNA to facilitate or prevent interaction of other factors 14 Principles of enhancer function: III. Interaction with the basal transcription apparatus Activators may directly interact with the basal transcription apparatus; often via a special domain - the activation domain (AD) Activators can also interact indirectly via separate factors, so called coactivators The same is true for repressors: Direct interaction via repressor domains or interaction via corepressors 15 The mediator complex links transcriptional regulators with the basal transcription apparatus Pol II Mediator is required for transcription from most Pol II dependent promoters in yeast – sometimes referred to as being a GTF itself 16 Alberts 2002 Nature Structural & Molecular Biology 11, 394 - 403 (2004) The mediator complex links transcriptional regulators with the basal transcription apparatus The modular structure of mediator allows interaction with different transcription factors, coactivators and components of the basal transcription apparatus (Cartoon! Not all interactions present at the same promoter....) 17 Nature Structural & Molecular Biology 11, 394 - 403 (2004) Current Opinion in Genetics & Development 18:397–403 (2008) Integration at promoters Promoters can function as genetic switches that integrate regulatory information MODULARITY of regulatory input is a recurring theme 18 Alberts 2002 Summary eukaryotic transcriptional regulation principles 1. Enhancer: activating regulatory sequence separate from core promoter – independent from distance and orientation 2. Enhancers bind activating transcription regulators (repressing factors bind to silencers) 3. Enhancers may function – in making the promoter accessible (chromatin remodelling and modifications) – changing DNA topology (e.g.bending) – interacting with the basal transcription apparatus 4. Promoters integrate information from various regulatory elements (modularity) More about all this in Clemens Grabher‘s lecture 19 RNA pol II promoter Nature. 2009 Sep 10;461(7261):186-92 Modular: can contain e.g. Inr – Initiator region TATA box or DPE – Downstream Promoter Element (in TATA-less promoters) TATAA YYCAYYYY AGAC reminiscent of prokaryotes: TTGACA TATAAT 20 The core promoter is bound by general (or „basal“) transcription factors (GTFs) A promoter recognition factor binds the promoter: TFIID (Transcription Factor for RNA pol II D) Consistst of many subunits: TBP – TATA Binding Protein TAFs – TBP Associated Factors idealized cartoon: subunit composition varies a lot – different TFIIDs recognize different promoters 21 Nat Rev Genet. 2010 Aug;11(8):549-58 TBP bends DNA at the TATA box Widens the minor groove Brings proteins binding to the promoter into closer proximity In some complexes, TBP is present but does not bind DNA 22 The different core promoter types are bound by different promoter recognition factors Müller F et al. J. Biol. Chem. 2007;282:14685-14689 Differential expression of core promoter recognition factors may contribute to cell type specific transcription regulation CpG islands – a hallmark of „housekeeping genes“23 CpG islands • found in housekeeping genes: constitutively expressed genes • increased density of the dinucleotide CG at the 5‘ end • CpGs less frequent in the rest of the genome – the Cs get methylated by DNA–methyl-transferases – then frequently disappear – why? methylation spontaneous deamination • Mutation to T 24 Stryer 2002 CpG islands In active promoters, DNA should be demethylated Promoters active in the germline are spared of methylation -> less mutation of C to T 25 Alberts 2002 (How can methylation be detected?) Some restriction enzymes are sensitive to methylation of their recognition sites 26 Summary eukaryotic promoters 1. 2. 3. 4. 5. 6. Modular (as in prokaryotes) Frequent motifs: TATA, Inr, DPE Various classes of promoters combine different motifs Promoter recognition complexes bind the promoters Classic example: TFIID (TBP and TAFs) Different recognition complexes binding different promoter classes may contribute to cell type specific regulation of transcription 7. CpG islands are a feature of housekeeping genes and reflect the demethylated state of their promoter DNA 27 Assembly of the basal transcription apparatus = After the binding of TFIID, other TFIIs and the polymerase itself bind: initiation complex transforms into elongation complex 28 First assembly steps TFIIA: - TFDII can bind to region extending farther upstream TFIIB: - binds adjacent to TBP (BRE - B Recognition Element) - determines promoter polarity - recruits the polymerase TFIIF: - binds polymerase - facilitates recruitment 29 Eukaryotic RNA polymerases Cartoon of protein gel from yeast RNA polymerase II: The bacterial subunits: especially catalytic units conserved no sigma – role fulfilled by the GTFs enzyme alone can transcribe, but not initiate 30 (archaeal and eukaryotic RNA polymerases are related) 31 Trends in Microbiology 6/6, 222-228 (1998) Open complex formation and promoter clearance TFIIE: - facilitates formation of initiation-competent polymerase - recruits TFIIH TFIIH: - multiple enzymatic activities - helicase -> melting of the DNA CTD-domain of the RNA Pol II gets phosphorylated – leaves promoter and starts to elongate 32 Phosphorylation of proteins is an effcient way of regulation The reaction is catalysed by protein kinases, which are target selective Phosphorylation may : - cause conformational changes - create or abolish binding sites for other proteins Phosphate groups may be removed by selective phosphatases 33 Stryer 2002 Phosphorylation of the CTD regulates transcription The heptad repeat: (YSPTSPS)n=26-52 Ser2 Ser5 Mediator binds unphosporylated CTD 9, 810-815 (October 2008) Alberts 2002 CTD TFIIH phosphorylates Ser5 -> promoter clearance P-TEFb phosphorylates Ser2 -> escape from pausing 34 Stalled transcription –promoter-proximal pausing Polymerase on Drosophila heat shock genes stalled 50 bp downstream of the TSS Released by P-TEFb: -> phosphorylates pausing associated factors and the CTD-Ser2 -> proper elongation 35 Chromosoma (2009) 118:1–10 Regulation by transcription factors at different steps may save different purposes Pho4: chromatin opening HSF: escape from pausing P-TEFb • tight control via promoter accessibility and subsequent initiation steps • can be relatively slow (example: acid phosphatase gene should only be induced when the cell needs phosphate) • rapid activation of paused polymerase • control may be leaky (example: heat shock genes need to respond rapidly to heat stress) Nature. 2009 Sep 10;461(7261):186-92 HSF The CTD code A CTD code for different phases of transcription 37 Nat.Rev.Gen.10:457-466 (2009) CTD code and integration with RNA processing the „cap“ Stryer 2002 2008, 20:260–265 More about RNA modifications in Harald König‘s lecture 38 Semi Cell & Dev Biol 18 (2007) 691–697 The „transcription factory“ model a cell nucleus stained for phosphylated Pol II (red) Polymerases are localized and thread DNA through the „factory“ Nat Rev Genet. 2009 Jul;10(7):457-66. Such factories can also be associated with zones enriched for splicing factors 39 („nuclear speckles“) Transcription and DNA repair Transcription and genome integrity affect each other, e.g. DNA lesions inhibit progress of the polymerase -> repair TFIIH participates in both processes (Human disease genes: Xeroderma pigmentosum: e.g. XPB, XPD Cockayne syndrome: CSA, CSB) Also: Transcription can affect mutagenesis or recombination rates More on DNA repair in Felix Loosli‘s lecture 40 Summary pol II transcription 1. The initiation complex assembles at the site of core promoter recognition factor binding 2. The TFIIH helicase function assists in promoter melting 3. The TFIIH kinase function phosphorylates the CTD domain of the polymerase (Ser5) – promoter escape 4. Many genes have paused polymerases near their 5‘ end 5. PTEFb kinase phosphorylates the CTD (Ser2) – productive elongation 6. CTD is differentially phosphorylated throughout the transcription cycle – the CTD code 7. Transcription and RNA processing are integrated – mediated by CTD code 8. Transcription factories - spatial organization of transcription in the nucleus 9. Transcription and DNA repair are linked (TFIIH) 41 Cell 133, May 16, 2008 (Sequential vs. holoenzyme assembly) 42 Critical Reviews in Biochemistry and Molecular Biology, 41:105–178, 2006 5 minutes break ! 43 The other polymerases Ribosomal subunits are assembled in the nucleolus Ribosomes consist of proteins and RNA (more in Felix and Clemens‘ lectures!) Alberts 2002 Which polymerases are required for ribosome synthesis? Pol I Pol III Pol II Pol I and III can constitute up to 80% of all transcription in rapidly growing cells! 44 Cooper 2002 The other polymerases: Pol I „christmas tree“ transcription of tandem rDNA arrays Alberts 2002 Synergistic binding of UBF and SL1 to the promoter recruits PolI and associated factors 45 The other polymerases: Pol I Many genes involved in cancer regulate components of the Pol I machinery – Reflects the importance of ribosome synthesis for cell growth and proliferation 46 The other polymerases: Pol III Pol III transcribes a whole battery of small RNAs, the most abundant of which are tRNAs and 5S rRNA Dec;23(12):614-22 (2007) 47 The other polymerases: RNA pol III Dec;23(12):614-22 (2007) Basal RNA pol III promoter elements can be downstream of the transcription start site 48 The other polymerases All three classes of polymerases bind via commitment factors TBP is involved in initiation in all three classes (not always binding to the DNA) 49 The other polymerases organelles Human mitochondrial RNA polymerase Molecular Cell 24, 813–825, 2006 Bakteriophage T7 RNA polymerase (In chloroplasts more complicated: NEP – nuclear encoded, phage type PEP – plastid encoded, eubacterial type) Single subunit polymerases Alberts 2002 50 (T7 often used in the lab for in vitro transcription) Other polymerases: summary • 3 eukaryotic polymerases: - RNApol I: rRNA (18S, 28S) - RNA pol II: mRNAs, other small RNAs - RNA pol III: tRNAs, 5S rRNA, other small RNAs (+ organelle polymerases: - in mitochondria: phage-like - in chloroplasts: both phage and eubacterial type) • TBP required for promoter recruitment in all 3 51 Some examples of current research 52 Quantifying transcription of the genome: RNA seq http://en.wikipedia.org/wiki/File:RNA-Seq-alignment.png 53 Nature Methods Vol5 No7 ,JULY 2008 pp.621-28 Global mapping of transcription start sites CAGE technology: „Cap Analysis of Gene Expression“ 54 http://www.riken.go.jp/engn/r-world/research/lab/osc/genotech/images/01b.gif Different types of promoters detected with CAGE 55 Nat Rev Genet. 2007 Jun;8(6):424-36 Different types of promoters May be linked with different core promoter recognition factors Müller F et al. J. Biol. Chem. 2007;282:14685-14689 56 Large scale sequencing of transcripts • Mapping of start sites • Alternative transcripts • Other transcripts, also many from intergenic regions • „Pervasive transcription“ of the genome „Dark matter transcripts“ 57 „Dark matter“ transcripts? Based on RNA seq results as opposed to the tiling array method => When a new technology is introduced, be aware of artefacts! 58 „Dark matter“ transcripts? However, the story continues….: Many newly discovered types of transcripts are associated with promoters Just transcriptional „noise“? Or specific functions? 60 Nat Rev Genet. 2009 Dec;10(12):833-44 Transcription at enhancers? eRNAs = enhancer RNAS When the promoter is missing, RNA polymerase still sits at the enhancer, but no transcription occurs Function of all this? 61 Thanks for your attention! 62 References Pictures without reference are from Lewin‘s Genes X, © Jones and Barlett publishers, LLC (www.jbpub.com) Pictures with the following reference are from: Alberts 2002: Alberts, Molecular Biology of the Cell: http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=mboc4 Stryer 2002: Stryer, Biochemistry: http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=stryer Cooper 2002: Cooper, The Cell – A Molecular Approach: http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=cooper Knippers 1997: Rolf Knippers, Molekulare Genetik, 7. Auflage, Thieme 1997 63