Molecular Weight Determination
advertisement
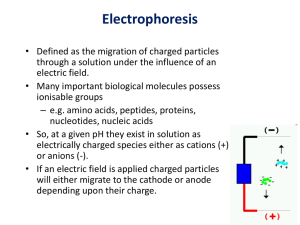
Molecular Weight Determination SDS-PAGE Electrophoresis • Movement of particles by an electric force. • Gel is the matrix perfused by buffer salts – Matrix will • reduce effects of diffusion • stabilize the system against turbulence (thermal agitation) • Introduce sieving action that allows separation based on molecular size – Buffer salts will • prevent sample interactions • create more conductivity • generates heat – Mobility = applied voltage x net charge on the molecule / function of molecule (shape, size and other physical matter) – Force: Total electric field is the effective charge and field strength – Opposing force is friction and pore size – When charge/mass is constant then large molecules move slower than small molecules. Electrophoresis • Two other factors are important – Ionic strength of surrounding solution • Buffer ions – – high ionic strength reduces effective charge of sample and temperature will increase; – low ionic strength slows the movement but has sharper peaks. • Best is Tris borate buffer – only few ions are free and large size ions reduce excessive heat generation – Temperature of the gel • Effects the viscosity of the gel • Need to run at constant power • SDS PAGE maintain high temperature to prevent renaturation Electrophoresis • Electric parameters: Power = I2R I = current R= resistance • Buffer ions: cations move towards negative electrode in upper chamber and anions move towards positive electrode in the lower chamber. • Apparatus and buffers provide constant resistance. Type of electrophoresis • Solution: Solutes dissolved in a solvent and electric force is applied. • Zonal: Aqueous ionic solution is carried in solid matrix. – Paper – cellulose acetate and cellulose nitrate – Gels • Starch gels – organelles • Agarose DNA • Polyacrylamide – proteins and nucleic acids Polymerization • Polyacrylamide gels are formed by the polymerization of acrylamide in aqueous solution in the presence of small amounts of a bifunctional crosslinker, usually bisacrylamide (bis). • The copolymerization of acrylamide with bisacrylamide produces a mesh-like network in three dimensions, consisting of acrylamide chains with interconnections formed from the bisacrylamide. Polyacrylamide gels • T -the total percentage concentration (w/v) of monomer (acrylamide plus crosslinker) in the gel. • C - the percentage of the total monomer represented by the crosslinker. • For example, an 8%, 19:1 (acrylamide/bisacrylamide) gel would have a T value of 8% and a C value of 5%. Polymerization • The polymerization of acrylamide and bisacrylamide occurs via a free-radical mechanism after adding a catalyst. • ammonium persulfate initiates free radicals and • tertiary aliphatic amine N,N,N',N'tetramethylethylenediamine (TEMED) is a catalyst for free radical propagation Polymerization • The presence of excess oxygen will inhibit the polymerization elongation process and can lead to shorter average chain length. • Deaeration under vacuum with a magnetic stirrer is needed prior to addition of initiators. Advantages of Polyacrylamide Gels • Small pore size. • By changing the concentration of the monomer and the cross-linker, the pore size can be varied in a reproducible manner. • It is synthetic so no batch to batch differences. SDS-PAGE • SDS (Sodium dodecyl sulfate) – PAGE (Poly Acrylamide Gel Electrophoresis) separation of proteins is based on molecular weight. • In their native form, proteins fold into a variety of shapes, some compact, some elongated. The rate of migration of native proteins through a sieving medium is therefore more a reflection of their relative compactness, and less an accurate measure of molecular weight. • Denaturing the proteins nullifies structural effects on mobility, allowing separation on a true charge/mass ratio basis. It also separates subunits in multimeric proteins, allowing analysis of large, complex aggregates. SDS • SDS is amphiphatic surfactant, an anionic detergent (pH above 7) • Hydrophobic tail binds to protein backbone • Polar heads denature ionic bonds • Uniform coating leading to charge/mass ratio consistent between proteins. 1.4g SDS/gram of protein is needed. SDS-PAGE • 2-mercaptoethanol – dissociates any disulfide bonds • SDS and urea disrupts ionic, hydrogen and hydrophobic bonds. • Overall – proteins have negative charge and disrupted secondary and tertiary charges. SDS-PAGE • Protein now have negative charge. When electric field is applied they move towards cathode. • Larger the size of the protein, the more it is retarded by contact with the meshwork of acrylamide in the gel, so the smallest peptides will move farther in the gel and the larger molecules move progressively shorter distances Discontinuous gel Electrophoresis • Two layers of different concentrations of gel and pH – Stacking low concentration and low pH – Separating gel – higher concentration of gel and high pH • Concentrate proteins in narrow band in stacking gel before traveling through separation gel for greater resolution of bands • Made up of stacking gel pH 6.8; separation gel, pH 8.8 and the tank . Discontinuous Gels • Stacking gel has low concentration of gel, so ions move freely under applied voltage. – Glycine and chloride ions are used • Chloride ions move faster (leading ions) than glycine (trailing ions) and create steep voltage gradient. Glycine ions are pulled due to the charge gradient and the two ions move at same speed. The proteins with intermediate mobility are carried along becoming stacked into very thin, distinctive layers in order of mobility. • Resolving gel has high concentration of gel and high pH: – When the interface between the stacking and separation gel is reached by moving, the pH changes abruptly. – Glycine moves quickly and unstacks the proteins. – The movement of proteins is influenced by sieving capacity of the gel and its molecular weight because pH and the voltage are constant. SDS-PAGE • After electrophoresis, the gel is treated with trichloroacetic acid solution – makes the proteins insoluble and – prevent them from diffusing during staining procedure SDS-PAGE • Staining to make invisible proteins visible. • Comassie blue in methanol-acetic acid Procedure • • • • Prepare Tank buffer 2x treatment buffer Staining solutions Destaining solutions I and II Protein Samples • Dissolve protein sample with 2x treatment buffer (protein conc. 50 µg). Boil for 5min to denature. • Standards: • Lactalbumin; trypsin inhibitor; trypsinogen; carbonic anhydrase; albumin egg; albumin bovine SDS-PAGE • Preparing standard curve – Rf = distance traveled by peptide / distance traveled by tracking gel • Plot a standard curve of log molecular weight Vs Rf