AssocWeirSection3
advertisement
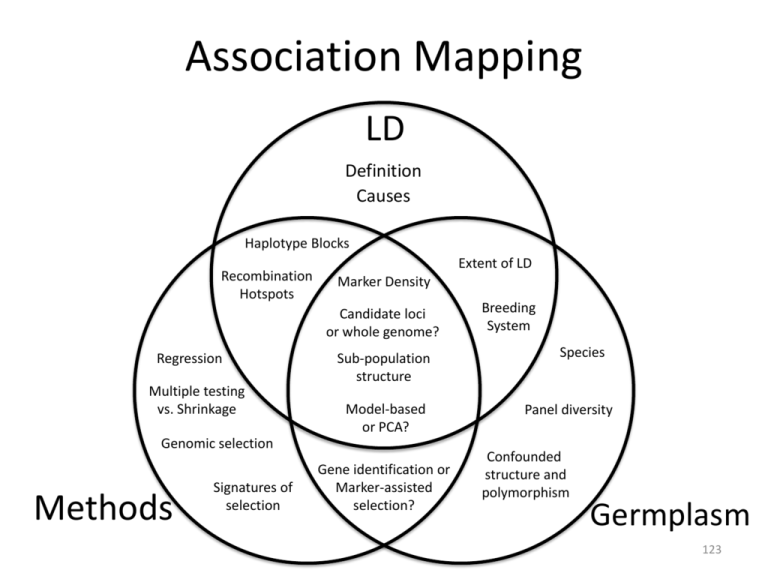
Association Mapping LD Definition Causes Haplotype Blocks Recombination Hotspots Extent of LD Marker Density Candidate loci or whole genome? Regression Multiple testing vs. Shrinkage Sub-population structure Model-based or PCA? Genomic selection Methods Signatures of selection Gene identification or Marker-assisted selection? Breeding System Species Panel diversity Confounded structure and polymorphism Germplasm 123 Outline • Association mapping is regression • Accounting for structure – Estimating structure using markers – Truly multi-factorial models • Miscelaneous topics: – Genomic control; TDT; Confounding with structure; Haplotype predictors; Genetic heterogeneity; Missing heritability; NAM; Validation 124 Association Mapping • It’s the same thing as linkage mapping in a biparental population but in a population that has not been carefully designed and generated experimentally • Because the experiment has not been designed, it is messy. Statistical methods are needed to deal with the mess 125 Regression • xi is the allelic state at a marker • Consider the total genotypic effect of I • qi is the allelic state at a QTL with which the marker is (hopefully) in LD • Now estimate β 126 Estimate of Beta 127 When is cov(x, g) non-zero? • Differences in allele frequencies at the marker between subpopulations AND difference in phenotypic mean between subpopulations – The difference in mean can be due to a single or many loci • Difference in the frequency of alleles between families AND difference in family phenotypic means within a (sub)population 128 Population structure Structure possibilities Familial relatedness Yu, J., Pressoir, G., et al. 2006. Nat Genet 38:203-208 129 Controlling for structure • Basic quantitative genetics: – Two individuals who share many alleles should resemble each other phenotypically – Use markers to figure out how many alleles individuals share and then use that to adjust statistically for their phenotypic resemblance 130 Controlling for structure • The “mixed model” Yu, J., Pressoir, G., et al. 2006. Nat Genet 38:203-208 131 Controlling for structure • Structure => large differences in allele frequencies across many markers Average marker Set2 score 1 0.8 First PCA axis 0.6 0.4 0.2 0 0 0.2 0.4 0.6 0.8 Average marker Set1 score 1 Regression coefficients of the phenotype on the PCA values 132 Use of PCA • Results are not sensitive to the number of PCA, provided you have enough – Price, A.L. et al. 2006. Principal components analysis corrects for stratification in genomewide association studies. Nat Genet 38:904-909 • The number of significant PC can be determined – Patterson, N. et al. 2006. Population Structure and Eigenanalysis. PLoS Genetics 2:e190 • Use a “Screeplot” 133 Historical footnote • PCA achieves what the Pritchard program Structure does • PCA is faster and more robust Pritchard, J.K. et al. 2000. Genetics 155:945-959 Price A.L. et al. 2006. Nat Genet 38:904-909. Patterson N. et al. 2006. PLoS Genetics 2:e190 134 Kinship • We are all a little bit related: – Two unrelated people: go back 1 generation, all four parents must be different people. – Go back 2 generations, all eight grand-parents must be different people. – Go back 30 generations, all 2.1 billion ancestors would need to be different people: Impossible! 135 Identity by Descent • Two alleles that are copies (through reproduction) of the same ancestral allele Coefficient of Coancestry • Choose a locus • Pick an allele from Ed and one from Peter • Probability that the alleles are IBD = Ed and Peter’s Coefficient of Coancestry, θEP 136 Coef. of Coancestry –> A matrix • A is the additive relationship or kinship matrix Winter Six-Row “Bison” Two-Row 137 A constrains u • Two individuals who share many alleles should resemble each other phenotypically • u is the polygenic effect • Its covariance matrix is Var(u) = Aσ2u • If aij has a high value, the ui and uj should have similar values (they have high covariance) • A constrains the values that are possible for u 138 Single locus, additive model: cov(ui, uj) 139 A matrix from the pedigree • The cells in the A matrix are aij = 2θij, the additive relationship coefficients between i in the row and j in the column • Coefficient of coancestry θij: the prob that a random alleles from i and j are IBD • Calculate from the pedigree by recursion: 140 A matrix from marker data , the homozygosities over all markers and alleles 141 With inbreeding, parental contributions NOT 50:50 • Maize intermated population • Drift during intermating and inbreeding • Markers can give more accurate θ than pedigree 90 80 70 60 50 40 30 20 10 0% 5% 10% 15% 20% 25% 30% 35% 40% 45% 50% 55% 60% 65% 70% 75% 80% 85% 90% 95% 100% 0 / 142 Mixed Model Example • • • • Five individuals, a, b, c, d, and e. a and b in subpop 1; c, d, and e in subpop2. a, b, c, and d unrelated; e is offspring of c and d. a and d carry the 0; b, c, and e carry the 1 allele y= μ + Xβ + Qv 143 Mixed Model Example • • • • Five individuals, a, b, c, d, and e. a and b in subpop 1; c, d, and e in subpop2. a, b, c, and d unrelated; e is offspring of c and d. a and d carry the 0; b, c, and e carry the 1 allele y= μ + Xβ + Qv 144 Mixed Model Example • • • • Five individuals, a, b, c, d, and e. a and b in subpop 1; c, d, and e in subpop2. a, b, c, and d unrelated; e is offspring of c and d. a and d carry the 0; b, c, and e carry the 1 allele y= μ + Xβ + Qv + Zu +e 145 Mixed Model Example • • • • Five individuals, a, b, c, d, and e. a and b in subpop 1; c, d, and e in subpop2. a, b, c, and d unrelated; e is offspring of c and d. a and d carry the 0; b, c, and e carry the 1 allele Zu A= var(u) = σ2u 146 Mixed Model Example • There is a polygenic effect u for each individual => overdetermined model? • NO: u is a random effect, constrained by Aσ2u 147 Mixed Model Example μ + Xβ + Qv y= + Zu +e –1 = ✕ 148 Control false positives from structure Flowering time (High population structure) Ear height (Moderate population structure) 0.5 0.5 0.5 a. 0.4 Cumulative P Ear diameter (Low population structure) b. Simple Simple 0.4 Q 0.3 c. 0.4 Q Q K GC Q+K 0.3 0.3 Q+K Simple K 0.2 K 0.2 0.2 Q+K GC 0.1 0.1 GC 0 0.1 0 0 0.1 0.2 0.3 Observed P 0.4 0.5 Simple Q K Q+K GC 0 0 0.1 0.2 0.3 Observed P 0.4 0.5 0 0.1 0.2 0.3 Observed P 0.4 0.5 A straight diagonal line indicates an appropriate control of false positives. Q + K model has best Type I error control, most important when trait is related to population structure (e.g., flowering time). 149 Statistical power Flowering time (High population structure) 1 Ear height (Moderate population structure) 1 d. 1 e. K Q+K Adjusted average power Ear diameter (Low population structure) Q+K 0.8 K Simple Simple Simple 0.6 K GC 0.4 0.2 0.2 0 0.4 (3.3) 0.6 (7.1) (11.9) Genetic effect (Phenotypic variation explained in %) 0.8 (17.4) 1 Simple Q K Q+K GC 0.2 0 0.2 (0.8) GC 0.4 0.4 GC 0 (0) Q+K Q 0.6 0.6 Q 0.8 0.8 Q f. 0 0 (0) 0.2 (0.8) 0.4 (3.3) 0.6 (7.1) (11.9) Genetic effect (Phenotypic variation explained in %) 0.8 (17.4) 1 0 (0) 0.2 (0.8) 0.4 (3.3) (7.1) 0.6 (11.9) 0.8 (17.4) Genetic effect (Phenotypic variation explained in %) Q + K model had highest power to detect SNPs with true effects. 150 1 Controlling for Structure Original P Matrix K Matrix 151 FDR vs. Power for 300 lines, 10 QTL 152 Effect of line number, P-only 10 QTL, 0.75 heritability 153 Effect of Reduced Population Diversity 154 Take homes on diversity • At equal population size – A less diverse population can increase power because relative to the extent of LD, the average marker distance is lower – Given that you are testing fewer markers, the multiple testing problem is reduced • Avoid as much as possible reducing population size for the sake of obtaining a more homogeneous population 155 Guidelines • • • • More lines and more markers are better For a diverse population, 800+ lines For a narrower population, 300+ (?) FDR is a reasonable method of determining significance, but probably conservative 156 1680 2360 1330 2320 1290 2280 1250 Q constant K estimated 1640 1600 • Q estimated with all markers, K estimated with varying fraction of markers available 1560 2240 1210 1520 2200 1170 1480 2160 0% 25% 50% 75% 100% 1130 0% 25% Flowering time Variance ratio d 50% 75% 100% 0% Ear height e 0.80 f 0.80 0.60 0.40 0.40 0.40 0.20 0.20 0.20 0.00 25% 50% 75% Marker number 100% 75% 100% 0.80 0.60 0% 50% Ear diameter 0.60 0.00 25% SSR SNP 0.00 0% 25% 50% 75% Marker number 100% 0% 25% 50% 75% 100% Marker number 157 1680 2360 1330 2320 1290 2280 1250 Q estimated K constant 1640 1600 • Q estimated with varying fraction of markers available, K estimated with all markers 1560 2240 1210 1520 2200 1170 1480 2160 0% 25% 50% 75% 100% 1130 0% Flowering time Variance ratio d 25% 50% 75% 100% 0% Ear height e 0.80 f 0.80 0.60 0.40 0.40 0.40 0.20 0.20 0.20 0.00 25% 50% 75% Marker number 100% 75% 100% 0.80 0.60 0% 50% Ear diameter 0.60 0.00 25% SSR SNP 0.00 0% 25% 50% 75% Marker number 100% 0% 25% 50% 75% 100% Marker number 158 History / future of controlling for structure 159 Single locus: model mis-specification • “the problem is better thought of as model mis-specification: when we carry out GWA analysis using a single SNP at a time, we are in effect modeling a multifactorial trait as if it were due to a single locus” – Atwell S. et al. 2010. Nature 465:627-631 160 History: Candidate locus studies • AM started out with candidate locus studies where the effects of few loci could be fitted • The biotechnology was not there to type more than a few loci • The genetic background needed to be accounted for somehow (see above) • In any event, the computational power was not there to fit all 106 loci simultaneously 161 Logsdon B. et al. 2010. BMC Bioinformatics 11:58. Future: GWAS fitting all loci • These methods could displace mixed models accounting for structure 162 Sundry topics • • • • • • • Other methods to control structure QTL confounded with structure Single markers or haplotypes? Genetic heterogeneity Missing heritability Linkage disequilibrium / Linkage analysis Validation 163 Genomic Control • Calculate bias in distribution of test statistic using “neutral” loci, then account for bias • Devlin, B. and Roeder, K. 1999. Genomic Control for Association Studies. Biometrics 55:997-1004. • Works best for candidate genes: test loci can be distinguished from neutral control loci. Works less well for whole genome scans • • • Marchini, J. et al. 2004. Nat. Genet. 36:512-517 Devlin, B. et al. 2004. Nat. Genet. 36:1129-1131. Marchini, J. et al. 2004. Nat. Genet. 36:1131-1131 164 Transmission Disequilibrium Test • Experimental rather than statistical control of effects of structure • Originally conceived for dichotomous (e.g., disease / no disease) traits • Affected offspring and both parents, of which one must be heterozygous • Test whether the a putative causal allele is transmitted more often that 50% of the time • Spielman, R.S. et al. 1993. Am. J. Hum. Genet. 52:506-516 165 TDT • Extensions for quantitative traits • Allison, D.B. 1997. Am. J. Hum. Genet. 60:676-690 • Extensions for larger-than-trio pedigrees • Monks, S.A., and N.L. Kaplan. 2000. Am J Hum Genet 66:576-92 • Using for populations under artificial selection • Bink, M.C.A.M. et al. 2000. Genetical Res. 75:115-121 166 QTL confounded with structure • Particularly important for QTL affecting adaptation, e.g., flowering time Camus-Kulandaivelu, L. et al. 2006. Genetics 172:2449–2463 167 Also in rice… Ghd7-0a Non-functional Ghd7-2 Weak allele Ghd7-0 Deleted Ghd7-1, Ghd7-3 Functional Given geographic distribution and role in adaptation, selection using this locus will have marginal utility Xue, W. et al. 2008. Nat Genet 40:761-767 168 Confounded QTL with structure • Association analysis will have difficulty identifying such QTL: the QTL needs to be polymorphic within subpopulations • Traditional linkage studies of crosses between members of different subpopulations should be very effective in this case • e.g., Xue, W. et al. 2008. Nat Genet 40:761-767 • Multi-factorial methods will have difficulty identifying loci under strong structure 169 Dwarf8: Confounded with structure • Thornsberry, J.M. et al. 2001. Nat. Genet. 28:286-289 – First structured association test applied to plants Camus-Kulandaivelu, L. et al. 2006. Genetics 172:2449–2463 170 Single markers or haplotypes? • The jury is still out • Infinite ways to simulate and analyze – Ne, QTL MAF, QTL effect, quantitative vs. binary, age of mutation • Ex. 1: Dramatically more power for haplotypes vs single markers • Durrant, C. et al. 2004. Am J Hum Genet 75:35-43 171 Single markers or haplotypes? • Ex. 2: Similar or lower power for haplotype method relative to single marker method • Zhao, H.H. et al. 2007. Genetics 175:1975-1986 • Process to sort out what method most appropriate for when still has to happen 172 Exploiting Haplotype Blocks • Objective: reduce the genotyping cost while capturing polymorphism at all (most) loci • Haplotype: series of alleles at adjacent polymorphic loci • Blocks: majority of diversity in few haplotypes • => Strong LD between loci within blocks; weak LD between loci across blocks • Knowledge of the allele at one locus provides much information on the alleles at other loci 173 What causes blocks? • Recombination heterogeneity: coldspots within blocks, hotspots between blocks • Random sampling of alleles and timing of mutation relative to recombination events 174 Evidence for mechanisms • Humans: High marker density resources – Observed LD structure reproduced best if recombination hotspots every ~ 100 kbp • Reich, D.E. et al. 2002. Nat Genet 32:135-142. Wall, J.D., and J.K. Pritchard. 2003. Am J Hum Genet 73:50215. – Block boundaries correspond with positions of high current recombination • Jeffreys, A.J. et al. 2005. Nat Genet 37:601-606 – Block boundaries consistent across different human populations • De La Vega, F.M. et al. 2005. Genome Res. 15:454-462 175 Hotspots exist in plants (Arabidopsis) too 70 kbp Kim, S. et al. 2007. Nat Genet 39:1151-1155 176 Blocks also arise randomly • No relation between historic recombination (histogram) and block boundaries (dark bars) • Verhoeven, K.J.F. and K.L. Simonsen. 2005. Mol. Biol. Evol. 22:735-740 177 Blocks in barley 40 ?? Mbp 178 Block cause matters • If blocks arise from a recombination process, they will be consistent across populations • Markers that tag blocks identified in one population will therefore be useful in others • If blocks arise from random processes, tags useful in one population will not be so in another 179 Haplotypes for discovery in barley • 2198 mapped SNP in 1807 lines across barley • Five methods – Traditional single SNP – Four gamete: use D’ to determine boundaries – Tree scan: single df contrasts based on parsimony – HapBlock: group to capture diversity – Sliding window of 3 SNP • Simulation: mask a SNP and pretend it’s a QTL • Real data: heading date on 1040 lines 180 Results • Simulation: single SNP best in 5 / 8 cases and never worse than the best haplotype method • CAUTION: the QTL had the same properties as the SNP: ideal for single SNP discovery • IF QTL simulated as recent mutations on blocks with haplotype properties THEN haplotype methods had higher power – Even then, single SNP did pretty well 181 Real data (heading date) Only Tree Scan All methods Only 4gamete 1H 2H 3H 4H 5H 6H 7H Chromosome 182 4gamete success • Rare recombinants split off early-heading lines 001 -0.99 3 000 -0.53 64 111 -0.69 12 010 -0.44 82 110 -0.48 386 * 011 0.16 489 183 Take-homes • Simulations don’t support use of haplotype blocks – But we don’t know how to simulate the true nature of QTL • With real data, a diversity of approaches might produce the most useful candidate list 184 Block vs tag identification • Blocks require position, tags do not • General tag marker approach: – Identify markers in high LD with each other – Retain only one – Aggressive: among tags, see if combinations can be used instead of single tags de Bakker, P.I.W. et al. 2005. Nat Genet 37:12171223 185 Reducing marker numbers • Tag SNP and Imputation: • Tag marker approach – Identify markers in high LD with each other – Retain only one – Aggressive: among tags, see if combinations can be used instead of single tags 186 Tagging works • Power is maintained; genotyping is reduced Power •Greedy •Best N •Random Tags •No LD Average marker spacing (kbp) de Bakker, P.I.W. et al. 2005 187 Tags serve as a base for imputation • Model-based imputation using fastPHASE Scheet and Stephens. 188 2006 Imputation on tag markers works 189 Jannink et al. 2007 Imputation can increase power Chromosomal Position (Mb) 190 Marchini et al. 2007 Imputation can increase power 191 Guan and Stephens 2008 Genome scans with low LD • Numerous species have too low LD to perform (as of yet) whole genome scans – You would need too many SNP on too many genos • “Nested Association Mapping” • Known as “Linkage disequilibrium linkage analysis” (LDLA) in animal genetics Meuwissen, T.H.E. et al. 2002. Genetics 161:373-379. Yu, J. et al. 2008. Genetics 178:539-551 192 NAM Design • B73 is the reference parent, crossed to 26 other inbred lines, representing a large part of maize diversity B73 CML52 26 Times … B73 F1 F1 RIL1 P39 RIL2 … RIL199 RIL200 RIL1 RIL2 … RIL199 RIL200 193 SSD RIL × Tzi8 Tx303 P39 Oh7B Oh43 NC358 NC350 MS71 Mo18W M37W M162W Ky21 Ki3 Ki11 Il14H Hp301 CML69 CML52 CML333 CML322 CML277 CML247 CML228 CML103 B97 25 DL B73 F1s 1 2 200 194 NAM Genotyping • Type parents at high density (2.5 M SNP…) • Type RIL at low density (10 k SNP): know, on a sub-cM scale, which parental allele inherited 195 NAM linear models • P: matrix indicating which parent contributed the allele to each offspring. α: vector of effects of parental alleles. • Eq. 1 • A linear model for the parental allele effects: • Eq. 2 • This latter model is what Yu et al. call “Projecting parental SNP on to the progeny.” 196 NAM / LDLA on Maize • Consider: – 2.5 Gbp genome with LD extending to 1000 bp – Requires 2.5 M SNP… • Apply Eq. 1 to identifiy QTL with, say, 3 cM C.I. – Within interval there are ~ 3000 parental SNP • Apply Eq. 2 to dissect the QTL to its causal SNP – Feasible with 25 genotypes (apparently) – Note that α will be accurately estimated 197 Advantages of NAM / LDLA • Adds power without adding huge genotyping burden • Reduces / eliminates problems related to structure: the linkage part of the analysis removes long-distance LD 198 Sugary1: Genetic heterogeneity Tracy, W.F. et al. 2006. Crop Sci 46:S-49-54 199 Genetic heterogeneity hinders AM • Distinct mutations at the B locus B2 associated with A1 B3 associated with A2 A1B1 Exists A2B1 Exists A1B1 Exists A2B1 Exists A1B2 New A2B2 A1B3 A2B3 New -- -- • If B2 and B3 cause a phenotype (e.g. loss of function at isoamylase), it will be associated with A1 in one case and A2 in the other case. • B2 and B3 can be identified by linkage mapping 200 How prevalent is heterogeneity? Buckler et al. 2009. Science 325:714-718 201 Multiple hits => Allelic series Buckler et al. 2009. Science 325:714-718 202 Heterogeneity and Pop. History • If a population has gone through a severe bottleneck, polymorphic loci are unlikely to have > 2 alleles… • Heterogeneity is less likely in domesticated populations with low Ne 203 204 Missing Heritability • Heritabtility for height in humans is ~ 0.80. • Very large GWAS studies find ~ 50 SNP together accounting for 5% of that heritability • Where’s the rest? – Infinitesimal effects – Low frequency SNP in same causal genes – Epigenetics – Genotype x environment interaction – Epistasis Maher, B. 2008. Nature 456:18-21 Manolio T.A. et al. 2009.Nature 461:747-753. 205 Plants are not like humans • Atwell et al. 2010. Nature 465:627-631 – Just 192 lines! – Some large effect variants (intermediate frequency and explain 20% of variation…) – Inbred lines enable noise reduction – Extended association peaks because of low Ne • Less evidence of missing heritability 206 Mouse composite not like humans • Valdar et al. 2006. Nature Genetics 38:879887 • QTL account for 73% of observed heritability 207 Humans are not like humans • Yang et al. 2010. Nat. Genet. 10.1038/ng.608 – Common SNP accounted for 45% of variation if all SNP included in the model • i. Many very small QTL effects • ii. QTL generally have lower MAF than arrayed SNP • Dickson S.P. et al. 2010. PLoS Biol 8:e1000294 – Several rare variants can combine to produce an association with a common SNP 208 Validation • All genome-wide studies raise the question of validation • In candidate studies, independent evidence from biological reasoning for candidate choice • In Zhao et al. 2007, used previous linkage analyses of parents in the association panel 209 Real data (heading date) Linkage Studies VRN3 1H 2H 3H 4H 5H 6H 7H Chromosome 210 Arabidopsis: Residual structure Residual Confounding: no bi-parental QTL found despite it segregating in the cross Low Power: No association found despite large effect in the cross 211 Recap • Model has focused on one locus at a time • The locus has been treated as a fixed effect – Makes sense in the candidate locus context • We have dealt with residual “polygenic” effects that, through structure, wreak havoc • Going forward, statistical models will be multi-factorial • Linkage mapping needed to find loci associated with structure • LD exhibits block-like structure: what to do with that? • Potential for genetic heterogeneity depends on population history • GWAS can miss substantial heritability • If you have very low LD, nested association, or LDLA, is a good idea 212







