S i-1
advertisement
Hidden Markov models in
Computational Biology
DTC
Gerton Lunter, WTCHG
February 10, 2010
Overview
First part:
Mathematical context: Bayesian Networks
Markov models
Hidden Markov models
Second part:
Worked example: the occasionally crooked casino
Two applications in computational biology
Third part:
Practical 0: a bit more theory on HMMs
Practical I-V: theory, implementation, biology. Pick & choose.
Part I
HMMs in (mathematical) context
Probabilistic models
Mathematical model describing how variables occur together.
Three type of variables are distinguished:
Observed variables
Latent (hidden) variables
Parameters
Latent variables often are the quantities of interest, and can be inferred
from observations using the model. Sometimes they are “nuisance
variables”, and used to correctly describe the relationships in the data.
Example: P(clouds, sprinkler_used, rain, wet_grass)
Some notation
P(X,Y,Z):
probability of (X,Y,Z) occurring
(simultaneously)
P(X,Y):
probability of (X,Y) occurring.
P(X,Y|Z): probability of (X,Y) occurring, provided that it is
known that Z occurs (“conditional on Z”, or “given Z”)
P(X,Y) = ΣZ P(X,Y,Z)
P(Z) = ΣX,Y P(X,Y,Z)
P(X,Y| Z ) = P(X,Y,Z) / P(Z)
ΣX,Y,Z P(X,Y,Z) = 1
P(Y | X ) = P(X | Y) P(Y) / P(X)
(Bayes’ rule)
Independence
Two variables X,Y are independent if P(X,Y) = P(X)P(Y)
Knowing that two variables are independent reduces the
model complexity. Suppose X,Y each take N possible values:
specification of P(X,Y) requires N2-1 numbers
specification of P(X), P(Y) requires 2N-2 numbers.
Two variables X,Y are conditionally independent (given Z) if
P(X,Y|Z) = P(X|Z)P(Y|Z).
Probabilistic model: example
P(Clouds, Sprinkler, Rain, WetGrass) =
P(Clouds) × P(Sprinker|Clouds) × P(Rain|Clouds)
× P(WetGrass | Sprinkler, Rain)
This specification of the model determines which variables are
deemed to be (conditionally) independent. These independence
assumptions simplify the model.
Using formulas as above to describe the independence relationship
is not very intuitive, particularly for large models. Graphical
models (in particular, Bayesian Networks) are a more intuitive way
to do the same
Bayesian network: example
Cloudy
P(Clouds) ×
P(Sprinker|Clouds) ×
P(Rain|Clouds) ×
P(WetGrass | Sprinkler, Rain)
Rule:
Sprinkl
er
Rain
Wet
grass
Two nodes of the graph are
conditionally independent given
the state of their parents
E.g. Sprinker and Rain are
independent given Cloudy
Bayesian network: example
Cloudy
P(Clouds) ×
P(Sprinker|Clouds) ×
P(Rain|Clouds) ×
P(WetGrass | Sprinkler, Rain)
Convention:
Sprinkl
er
Rain
Wet
grass
Latent variables are open
Observed variables are shaded
Bayesian network: example
Combat Air Identification algorithm; www.wagner.com
Bayesian networks
Intuitive formalism to develop models
Algorithms to learn parameters from training data
(maximum likelihood; EM)
General and efficient algorithms to infer latent variables from
observations (“message passing algorithm”)
Allows dealing with missing data in a robust and coherent way
(make relevant node a latent variable)
Simulate data
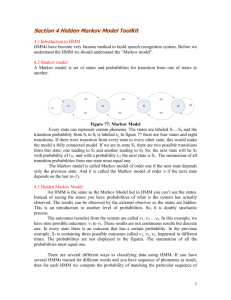
Markov model
A particular kind of Bayesian network
S1
S2
S3
S4
S5
S6
S7
S8
…
All variables are observed
Good for modeling dependencies within sequences
P(Sn | S1,S2,…,Sn-1 ) = P(Sn | Sn-1 ) (Markov property)
P(S1, S2, S3, …, Sn) = P(S1 ) P(S2|S1 ) … P (Sn | Sn-1 )
Markov model
States: letters in English words
Transitions: which letter follows which
S1
S2
S3
S4
S5
S6
S7
S8
…
MR SHERLOCK HOLMES WHO WAS USUALLY VERY LATE IN THE MORNINGS
SAVE UPON THOSE NOT INFREQUENT OCCASIONS WHEN HE WAS UP ALL ….
S1=M
S2=R
S3=<space>
S4=S
S5=H
….
P(Sn= y| Sn-1= x ) =
P(Sn-1Sn = xy ) / P (Sn-1 = x )
(frequency of xy) / (frequency of x)
(parameters)
(max likelihood)
UNOWANGED HE RULID THAND TROPONE AS ORTIUTORVE OD T HASOUT TIVE
IS MSHO CE BURKES HEST MASO TELEM TS OME SSTALE MISSTISE S TEWHERO
Markov model
States: triplets of letters
Transitions: which (overlapping) triplet follows which
S1
S2
S3
S4
S5
S6
S7
S8
…
MR SHERLOCK HOLMES WHO WAS USUALLY VERY LATE IN THE MORNINGS
SAVE UPON THOSE NOT INFREQUENT OCCASIONS WHEN HE WAS UP ALL ….
S1=MR<space>
S2=R<space>S
S3=<space>SH
S4=SHE
S5=HER
….
P(Sn= xyz| Sn-1= wxy ) =
P( wxyz ) / P( wxy )
(frequency of wxyz) / (frequency of wxy)
THERE THE YOU SOME OF FEELING WILL PREOCCUPATIENCE CREASON LITTLED
MASTIFF HENRY MALIGNATIVE LL HAVE MAY UPON IMPRESENT WARNESTLY
Markov model
States: word pairs
Text from: http://www.gutenberg.org/etext/1105
When thou thy sins enclose!
That tongue that tells the story of thy love
Ay fill it full with feasting on your sight
Book both my wilfulness and errors down
And on just proof surmise accumulate
Bring me within the level of your eyes
And in mine own when I of you beauteous and lovely
youth
When that churl death my bones with dust shall cover
And shalt by fortune once more re-survey
These poor rude lines of life thou art forced to break
a twofold truth
Hers by thy deeds
Then churls their thoughts (although their eyes
were kind)
To thy fair appearance lies
To side this title is impanelled
A quest of thoughts all tenants to the sober west
As those gold candles fixed in heaven's air
Let them say more that like of hearsay well
I will drink
Potions of eisel 'gainst my strong infection
No bitterness that I was false of heart
Though absence seemed my flame to qualify
As easy might I not free
Hidden Markov model
HMM = probabilistic observation of Markov chain
Another special kind of Bayesian network
S1
S2
S3
S4
S5
S6
S7
S8
…
y1
y2
y3
y4
y5
y6
y7
y8
…
Si form a Markov chain as before, but states are unobserved
Instead, yi (dependent on Si) are observed
Generative viewpoint: state Si “emits” symbol yi
yi do not form a Markov chain (= do not satisfy Markov property)
They exhibit more complex (and long-range) dependencies
Hidden Markov model
S1
S2
S3
S4
S5
S6
S7
S8
…
y1
y2
y3
y4
y5
y6
y7
y8
…
Notation above emphasizes relation to Bayesian networks
Different graph notation, emphasizing “transition probabilities”
P(Si|Si-1). E.g. in the case Si {A,B,C,D}:
Notes:
“Emission probabilities” P( yi | Si )
not explicitly represented
Advance from i to i+1 also implicit
Not all arrows need to be present (prob = 0)
A
B
C
D
Pair Hidden Markov model
y1
y2
y3
y4
y5
S11
S21
S31
S41
S51 …
S12
S22
S23
S24
S25 …
S31
S32
S33
S34
S35 …
z1
z2
z3
Pair Hidden Markov model
y2 in sequence y,
y3or in z, or both,
y4or neither (“silent”
y5
States y
may
1 “emit” a symbol
state).
… E.g.
S21 the associated
S31 coordinate Ssubscript
S51by one.
S11
If a symbol is emitted,
increases
41
diagonal transitions are associated to simultaneous emissions in both sequences.
A realization of the pair HMM consists of a state sequence, with each symbol
z1 emitted by exactly one state, and the associated path through the 2D table.
…
S
S
S
S
S
12
22
23
24
25
(A slightly more general viewpoint decouples the states and the path; then the hidden
z2
variables are the sequence of states S, and a path through the table. In this viewpoint
the transitions, not states, emit symbols. The technical term in finite state machine
theory is Mealy machine; the standard viewpoint is also known as Moore machine)
S31
S32
S33
S34
S35 …
Normalization:
Σpaths pΣsp(1) … sp(N)Σy1…yAΣ z1…zB P(sp(1),…,sp(N),y1…yA,z1…zB) = 1
z3
N = N(p) = length of path
Inference in HMMs
So HMMs can describe complex (temporal, spatial)
relationships in data. But how can we use the model?
A number of (efficient) inference algorithms exist for HMMs:
Viterbi algorithm: most likely state sequence, given observables
Forward algorithm: likelihood of model given observables
Backward algorithm: together with Forward, allows
computation of posterior probabilities
Baum-Welch algorithm: parameter estimation given observables
Summary of part I
Probabilistic models
Observed variables
Latent variables: of interest for inference, or nuisance variables
Parameters: obtained from training data, or prior knowledge
Bayesian networks
independence structure of model represented as a graph
Markov models
linear Bayesian network; all nodes observed
Hidden Markov models
observed layer, and hidden (latent) layer of nodes
efficient inference algorithm (Viterbi algorithm)
Pair Hidden Markov model
two observed sequences with interdependencies, determined by an unobserved
Markov sequence
Part II
Examples of HMMs
Detailed example:
The Occasionally Crooked Casino
Dirk Husmeier’s slides
http://www.bioss.sari.ac.uk/staff/dirk/talks/tutorial_hmm.pdf
Slides 1-15
Recommended reading:
Slides 16-23: the Forward and Backward algorithm, and
posteriors
Applications in computational biology
Dirk Husmeier’s slides:
http://www.bioss.sari.ac.uk/staff/dirk/talks/tutorial_hmm_bioinf.pdf
Slides 1-8: pairwise alignment
Slides 12-16: Profile HMMs
Part III
Practicals
Practical 0:
HMMs
What is the interpretation of the probability computed by the
Forward (FW) algorithm?
The Viterbi algorithm also computes a probability. How does
that relate to the one computed by the FW algorithm?
How do the probabilities computed by FW and Backward
algorithms compare?
Explain what a posterior is, either in the context of
alignment using an HMM, or of profile HMMs.
Why is the logarithm trick useful for the Viterbi algorithm?
Does the same trick work for the FW algorithm?
Practical I:
Profile HMMs in context
Practical I:
Profile HMMs in context
Lookup protein sequence of PRDM9 in the UCSC genome browser
Search Intropro for the protein sequence. Look at the ProSite profile and sequence logo.
Work out the syntax of the profile (HMMer syntax), and relate the logo and profile.
Which residues are highly conserved? What structural role do these play? Which are not
very much conserved? Can you infer that these are less important biologically?
Read PMID: 19997497 (PubMed). What is the meaning of the changed number of zinc
finger motifs across species? Relate the conserved and changeable positions in the zinc
fingers to the INTERPRO motif. Do these match the predicted pattern?
Read PMID: 19008249 and PMID:20044541. Explain the relationship between the
recombination motif and the zinc fingers. What do you think is the cellular function of
PRDM9?
Relate the fact that recombination hotspots in Chimpanzee do not coincide with those in
human with PRDM9. What do you predict about recombination hotspots in other
mammalian species? Why do you think PRDM9 evolves so fast?
Background information on motif finding:
www.bx.psu.edu/courses/bx-fall04/phmm.ppt
http://compbio.soe.ucsc.edu/SAM_T08/T08-query.html
Practical II:
HMMs and population genetics
Practical II:
HMMs and population genetics
Read PMID: 17319744, and PMID: 19581452
What is the difference between phylogeny and genealogy? What is
incomplete lineage sorting?
The model operates on multiple sequences. Is it a linear HMM, a
pair HMM, or something else?
What do the states represent? How could the model be improved?
Which patterns in the data is the model looking for? Would it be
possible to analyze these patterns without a probabilistic model?
(Estimate how frequently (per nucleotide) mutations occur
between the species considered. What is the average distance
between recombinations?)
How does the method scale to more species?
Practical III:
HMMs and alignment
Practical III:
HMMs and alignment
PMID: 18073381
What are the causes of inaccuracies in alignments?
Would a more accurate model of sequence evolution improve
alignments? Would this be a large improvement?
What is the practical limit (in terms of evolutionary distance, in
mutations/site) on pairwise alignment? Would multiple alignment
allow more divergent species to be aligned?
How does the complexity scale for multiple alignment using
HMMs, in a naïve implementation? What could you do to
improve this?
What is posterior decoding and how does it work? In what way
does it improve alignments, compared to Viterbi? Why is this?
Practical IV:
HMMs and conservation: phastCons
Practical IV:
HMMs and conservation: phastCons
Read PMID: 16024819
What is the difference between a phyloHMM and a “standard” HMM?
How does the model identify conserved regions? How is the model
helped by the use of multiple species? How is the model parameterized?
The paper uses the model to estimate the fraction of the human genome
that is conserved. How can this estimate be criticized?
Look at a few protein-coding genes, and their conservation across
mammalian species, using the UCSC genome browser. Is it always true
that (protein-coding) exons are well conserved? Can you see regions of
conservation outside of protein-coding exons? Do these observations
suggest that the model is inaccurate?
Read PMID: 19858363. Summarize the differences of approaches of the
new methods and the “old” phyloHMM.
Practical V:
Automatic code generation for HMMs
Practical V:
Automatic code generation for HMMs
http://www.well.ox.ac.uk/~gerton/Gulbenkian/HMMs and alignments.doc.
Skip sections 1-3.
Implementing the various algorithms for HMMs can be hard work, particularly
when a reasonable efficiency is required. Library implementations are however
neither fast nor flexible enough. This practical demonstrates a code generator
that takes the pain out of working with HMMs.
This practical takes you through an existing alignment HMM, and modifies it to
identify conserved regions (à la phastCons)
Requirements: a Linux system, with Java and GCC installed.
Experience with C and/or C++ is helpful for this tutorial.