TCR
advertisement

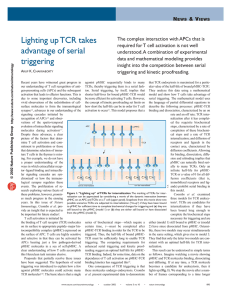
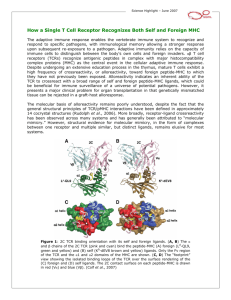
Team CDK Daniel Packer Rafael Rodriguez Sahat Yalkabov TCR Signaling Pathway TCR T (T cell receptor) Molecule found on T Cells Response for recognizing antigens on MHC (Major Histocompatibility Complex) cell is activated when TCR engages with antigen T cells Belong to white blood cells group called lymphocytes Play central role in immunity Distinguished from other cells by the presence of TCR on its surface Named T cells because they mature in thymus Electron micrograph of T cell T cell formation CD8+ and CD4+ T cells CD8+ (Cytotoxic T cells) Destroy virally infected cells and tumor cells Transplant rejection Recognize targets by binding to antigens associated with MHC class I Present on ~99.9% of the cells in the body Deactivated to anergic (inactive) state with the help of molecules secreted by the T-reg cells To prevent autoimmune diseases CD8+ and CD4+ T cells CD4+ (Helper T cells) Assist white blood cells with immunologic processes, as well as activation of cytotoxic T cells Activates with peptide antigens from MHC class II molecules (pMHC) Expressed on the surface of APCs When activated, divide rapidly and secrete small proteins called cytokines Regulate or assist active immune response MHC Cell surface molecule Mediate interactions between white blood cells and other immune cells or the body cells Determines compatibility of organ transplants Measures the susceptibility to autoimmune diseases In humans, MHC also called HLA (human leukocyte antigen) MHC region occurs on chromosome 6 Structure of TCR Member of immunoglobulin superfamily Consists of 2 halves: Alpha/Beta and Gamma/Delta fragments Structure similar to immunoglobulin Fab fragments Generation of TCR 1/2 Alpha/Gamma chain - generated by VJ recombination Beta/Delta chain – generated by V(D)J recombination Intersection corresponds to CDR3 region Important for antigen-MHC recognition Generation of TCR 2/2 Involves random joining of gene segments to complete TCR chain Unique combinations of segments, as well as palindromic and random N- and Pnucleotide additions accounts for great diversity T cell activation 1/2 TCR complex identifies specific bound antigen and elicits a distinct response The mechanism by which T cells evoke response is called T cell activation The most common mechanism for activation is via phos./dephos. by proten kinases. TCR associated reactions kinases: Lck Fyn CD45 Zap70 T cell activation 2/2 pMHC(agonist) Interacts even at low concentrations pMHC(endogenous) Weak interactions / No effect Target molecule: ERK Extracellular signal-regulated kinases Involved in regulation of meiosis, mitosis, and postmitotic. Activates on: Growth factors, cytokines, virus infection, transforming agents, carcinogens. Results Cumulative Distribution 1.00 0.90 0.80 Probability 0.70 pMHC(p~ag)=10 0.60 pMHC(p~ag)=100 0.50 pMHC(p~ag)=1000 0.40 pMHC(p~ag)=10k 0.30 pMHC(p~ag)=100k 0.20 No LCK Dephosphorilation 0.10 0.00 0.00 2.00 4.00 6.00 8.00 Time 10.00 12.00 14.00 Results pMHC(p~ag)=10 Probability Distribution 18 16 14 Frequency 12 10 8 pMHC(p~ag)=10 Frequency 6 4 2 0 6-7 7-8 8-9 9-10 10-1111-1212-1313-1414-1515-1616-1717-18 Time Results pMHC(p~ag)=100 Probability Distribution 18 16 Frequency 14 12 10 8 Frequency 6 4 2 0 Time Results pMHC(p~ag)=1000 Probability Distribution 40 35 Frequency 30 25 20 Frequency 15 10 5 0 7-8 8-9 9-10 10-11 Time 11-12 12-13 Results pMHC(p~ag)=10k 60.00 50.00 Frequency 40.00 30.00 Frequency 20.00 10.00 0.00 5-6 6-7 7-8 Time 8-9 Results pMHC(p~ag)=100k 70 60 Frequency 50 40 30 Frequency 20 10 0 4-5 5-6 Time 6-7 Conclusion pMHC(endogenous) had little to no effect on the activation times of ERK pMHC(agonist) had a very noticeable effect on the activation times of ERK ERK concentration starts around 202,000 and tops out at 296,000 The activation times of ERK depend on pMHC(agonist) concentrations The greater pMHC concentration, quicker are the activation times and smaller the time distribution