downloading - NeuralEnsemble
advertisement

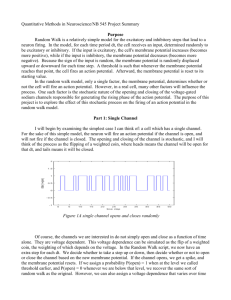
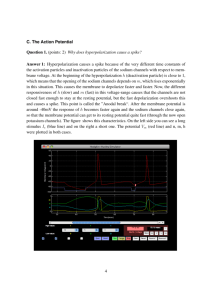
http://brian.di.ens.fr Dan Goodman & Romain Brette Ecole Normale Supérieure Projet Odyssée goodman@di.ens.fr brette@di.ens.fr Structure of a neuron Synapse Axon Dendrites Soma (cell body) Membrane potential Potential difference (V) The difference in concentrations of sodium and potassium ions (mostly). Roughly -70 mV at rest Inside cell Membrane (semi-permeable) Outside cell Action potentials (aka spikes) Synapses Neurotransmitter Presynaptic terminal Postsynaptic terminal Synaptic cleft Model neurons • • • • “Single compartment model” – the simplest Time evolution (differential equation) Spike propagation (delta function) Spike initiation – Threshold condition – Action potential (spike) – Reset V Integrate and Fire model • One variable V • No time evolution, or dV/dt=0 • Spike propagation, when spike arrives set V→V+w • Threshold, fire spike if V>Vt • Reset, after spike V→Vr Leaky I&F • One variable V • Exponential decay (‘leak current’) or τ dV/dt = -(V-Vr) • Spike propagation, when spike arrives set V→V+w • Threshold, fire spike if V>Vt • Reset, after spike V→Vr Introducing Brian – leaky I&F • Code is in Python • Equations (differential, define time evolution) • Threshold • Reset • Model • NeuronGroup • Connection Brian is flexible • Threshold increases when spike arrives and decays • Implemented as DE and user-defined reset and threshold functions Efficiency: vectorisation • Python is slow (interpreted) • In Brian most operations are vector operations (same operation on multiple pieces of data) • Use NumPy for vector operations • Linear differential equation, use matrix algebra for exact update t→t+dt • Spike propagation, V→V+w for certain V, w • Threshold, V>Vt • Reset V→Vr Data structures State matrix S, values of model variables at any given time V=0 x=1 y=2 Weight matrix W, synapse strengths Update matrix A Encodes exact solution to linear differential equation Brian’s vector operations Update matrix A State matrix S V x y Time evolution S=dot(A,S) * Threshold spikes=(S[0,:]>Vt).nonzero()[0] Spike propagation S[0,:]+=W[spikes,:] Reset S[0,spikes]=Vr The End • Brian is useful for modelling if: – Network of spiking neurons – Each neuron is modelled as single compartment – Not too many neurons (tens of thousands?) • Benefits are: – Easy to learn and use compared to other software – Quick to implement and tweak models