t=0
advertisement

August 13 2009
MOBIL Summer School
Lea Thøgersen
Model based on observations and theory.
Used to predict and explain new observations
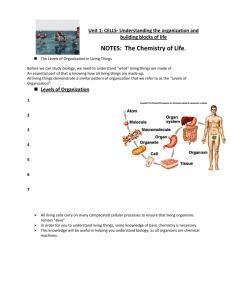
Molecular Modeling
Use the computer as a laboratory
Do you know any methods?
What are they used for?
Today: Molecular Dynamics
Experimental observations and simple physical
rules combined to simulate how different atoms
move wrt each other.
Topics:
Conformational energy, force field and molecular dynamics
Literature: “Part 3” (Chap. 8 Diffraction and Simulation)
p.196-200 (first 4 lines), p. 203-207, p. 210-212.
Goal: Obtain basic feeling for the possibilities and limitations
of molecular dynamics
?
Means: active participation from you
First session “Conformational Energy and Force Fields”
ends with an exercise
Second session “Molecular Dynamics”
includes discussion of a current research study
?
Etot = Ekin + Epot
Ekin for a molecule ?
?
e.g. vibration, diffusion
{½mv2}
coupled with temperature and atom velocities, but
independent of atom positions
Epot for molecule ?
{mgh (gravity) ; ½kx2 (spring)}
atoms affect each other dependent on atom type and
distance
=> Epot coupled with atom positions
conformational energy
C4H10
?
Atoms
?
nuclei (protons+neutrons)
electrons
Quantum Mechanics:
when chemical bonds are formed electrons redistribute on all atoms in the
molecule
a carbon (e.g.) would be different from molecule to molecule
the distribution of both the electrons and the nuclei in a molecule
determines the conformational energy
Experimentally:
atoms of particular type and in particular functional groups behave similar
independent of the molecule
IR wave lengths and NMR chemical shifts have characteristic values for certain
atom types and groups independent of which molecule they are a part of
Molecular Mechanics:
Conformational energy from distribution of only the nuclei
Not without problems
Energy as function of the relative positions of the atoms
=> conformational energy
Additive energy contributions
Spectroscopy of small molecules suggest that energy
contributions from individual internal coordinates are
independent, to a good approximation
Energy function as sum of independent contributions
Relative energies instead of absolutes
Easier to define energy penalty than absolute energy
Constant contributions can be ignored
E1 E1 S
E2 E2 S
E2 E1 E2 E1
Divided in “bonding” and “non-bonding” contributions
Describing the physics and chemistry of the atom interactions
bond stretch
angle bend
bond rotation
=> dihedral
E
E
eq
r or θ
Ebond
bonds i
1
2
φ
kibond ri req,i
2
angles i
1
2
kiangle i eq,i
2
dihedral i n
1
2
Vin 1 cos ni
Describing the physics and chemistry of the atom interactions
Electrostatic interactions
Van der Waals interactions
1500
Energy / kcal/mol
0.5
0.4
Energy
0.3
0.2
0.1
500
+
÷
0
2
4
0
2
4
6
8
6
8
0
-0.1
3
3.5
4 rij / Å 4.5
R
ij
rij
i j i
min
ij
12
5
5.5
R
2
rij
min
ij
6
qq
i j
i j i 4 0 rij
Energy / kcal/mol
0
Enon-bond
+
÷
1000
-500
-1000
-1500
+
÷
rij / Å
÷
+
Constants in the energy expression should be
determined
ex. E k r r ?
Based on experimental observations and QM
computations.
Hard and tedious work to construct a good
and general force field.
1
2
bond
bonds i
bond
i
2
i
eq,i
Gravity
Spring
Generally
Epot
mgh
?
||F||
mg
½k(Δx)
?2
Ep
k Δx
∂Ep/∂x
x
?
?
equilibrium
F=0
∆x > 0
F<0
∆x < 0
F>0
?
?
?
F?= - ∂Ep/∂x
Force Field
F Ep
Ep
R
The form of the potential
energy function defines a
force field
Function describing the potential energy of the molecule as a
function of atom positions - conformational energy
+Parameterization of this energy function
Examples: MMFF, CHARMM, OPLS, GROMOS…
Potential energy surface
Complex energy surface
Molecule specific
Only two out of 3N-6
variables shown here.
Minima correspond to
equilibrium structures
Q1 Bond Stretch: Which of the three lines represent the stretching of the
double C=C bond in propene and why?
Number 2. The equilibrium is found for a shorter distance (than for the solid line), and the
graph is steeper, meaning the force constant is higher, meaning the bond is stronger.
Q2 Bond Rotation: Which line represents the single bond, which
represents the double bond and why?
How many interactions contribute in fact to the rotation around the single and
the double bonds?
Number 1 = single bond, number 2 = double bond.
Number 1 has three minima (characteristic of an sp3 bond) and a low rotation barrier.
Number 2 has two minima (characteristic of an sp2 bond) and a high rotation barrier.
The double bond rotation has four contributions (5-1-2-6, 5-1-2-3, 4-1-2-6, 4-1-2-3)
The single bond rotation has six contributions (6-2-3-{7,8,9} and 1-2-3-{7,8,9})
Q3 vdW Interactions: Which line represents the H-H interaction, which
represents the C-H interaction and why?
Number 1 = H-H interaction, number 2 = C-H interaction. Hydrogen is a smaller atom than
carbon, and therefore the minimum vdW distance is smaller for H-H than for H-C.
Q4: What constitutes a force field, and why does it make sense to call it a ”force
field”? A force field consists of a potential energy function and the parameters for the function.
It is called a force field since the first derivative of the potential energy wrt the position
of an atom gives the force acting on this atom from the rest of the atoms in the system.
A Virtual Experiment
Both potential and
kinetic energy
Given a start structure
and a force field an MD
simulation output the
development of the
system over time
(nanosecond time scale)
2005
314,000 atoms
10 ns
1997
36,000 atoms
100 ps
LacI-DNA complex
ER DNAbinding domain
2007-8
1,000,000 atoms
14 ns
Satellite tobacco mosaic virus,
complete with protein, RNA, ions
ri(0)
vi(0)
ai(0)
ri(t)
vi(t)
ai(t)
?
ri(t+ δt)
vi(t+ δt)
ai(t+δt)
atom positions
atom velocities
atom accelerations
?
Time line
time step
∆t, δt
typical ∆t ≈ 1·10-15s = 1 fs
Find initial coordinates r(t=0)
for all atoms in the system
For proteins an X-ray or NMR structure is used or
modified
Water and lipid can be found pre-equilibrated
from the modeling software or on the web
Smaller molecules can be sketched naively and
pre-optimized within the modeling software
Avoid boundary effects
Every atom ’sees’ at
most one picture of the
other atoms.
Cutoff less than half the
shortest box side
At least 10Å cutoff.
Spring
Generally
Epot
½k(Δx)2
||F||
k Δx
Ep
∂Ep/∂x
x
equilibrium
F=0
∆x > 0
F<0
∆x < 0
F>0
F = - ∂Ep/∂x = -G
F=ma
r(t=0) => F(r(t=0)) => a(t=0)
?
Maxwell-Boltzmann distribution for kinetic
energy εk = ½mv2 => v(t=0)
Initial distribution of speed reproducing the
requested temperature
random directions
of the velocities
ri(t)
ri t t ri t t v t 12 t 2ai t
ai(t)
ai t t Fi t t mi m1 E p (r ) ri
vi(t)
vi t t vi t 12 t ai t t ai t
i
Time line
time step
∆t, δt
typical ∆t ≈ 1·10-15s = 1 fs
ri t t
Time line
time step
∆t, δt
typical ∆t ≈ 1·10-15s = 1 fs
Good
Collisions should occur smoothly!
Time step ~ 1/10 Tfast motion period
TC-H vib ~ 10 fs => Time step = 1 fs
?
Bad
?
Total simulation time e.g. 10 ns = 10.000.000 conformations
Build the system
Minimization of the system
Some 2000 steps, gradient < 5 or so
To remove clashes
Equilibration of the system
Clean pdb-structure for unwanted atoms
Add missing atoms
Add the environment
Make a structure file describing connections
Maybe constraining some atoms to their initial position too keep overall structure
Maybe starting from low temperature, and slowly increasing it to the wanted
Maybe letting the volume adjust properly to the size of the system
Energy and RMSD should level out
Production run
Constant temp, vol, pressure?
Experimenting with different setups to see what
happens – is the system stable?
Mutations, temperature, pressure, environment....
Test out hypotheses based on experiment
Detailed information at the atomic level
Free energy differences – site-directed mutagenesis
Other thermodynamics stuff
Poke it / steer it
X-ray, NMR and various biophysical studies and mutation
studies and more?
Model the hypothesis, does the modelled response fit the
experiment? If so, both the experiment and simulation conclusion is
strengthen and a higher level of understanding is gained
Shortcomings of MD:
Timescale - ns is very short – no conformational changes
System size – the dimensions of the model are less than nm
No electrons – polarization cannot be described
6 simulation setups. 10 ns simulations of SERCA in a membrane consisting of
either short, POPC, long, DMPC, or DOPC lipids, and SERCA in a membrane of
2:1 C12E8:POPC. 200-240.000 atoms.
X-ray low resolution scattering from bilayer leaflets. The bilayers in the crystals
consist of 16:7 detergent:lipid (detergent C12E8, lipids from native membrane).
Try to come up with relevant and interesting things to study
from the MD simulations.
POPC+detergent
POPC
Long
α
Membrane type
POPC:C12E8 (1:2)*
Short
DMPC
DOPC
POPC
Long
purePOPC
purePOPC:C12E8 (1:2)*
Avg. Hydrophobic thickness (8-10 ns) (Å)
< 7 Å from protein
> 7 Å from protein
26.3
26.9
27.0
29.4
29.5
32.7
23.3
27.0
28.4
30.7
30.7
34.3
31.3
24.0
Avg. Overall tilt
(8-10 ns) (°)
24.2
21.4
22.1
16.8
18.7
17.5
-
From Theoretical and Computational Biophysics Group,
University of Illinois at Urbana-Champaign
http://www.ks.uiuc.edu/Gallery/
K+ permeation
Voltage bias
Conduction via
knock-on mechanism
Selective filter
transmembrane pore of alphahemolysin
Electrophoretically-driven
58-nucleotide DNA strand
Full structure of
satellite tobacco
mosaic virus, complete
with protein, RNA,
ions, and a small water
box