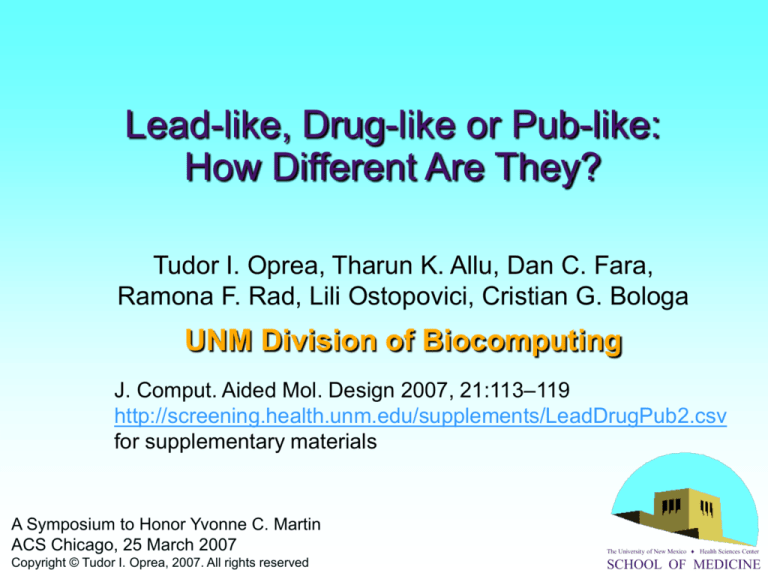
Lead-like, Drug-like or Pub-like:
How Different Are They?
Tudor I. Oprea, Tharun K. Allu, Dan C. Fara,
Ramona F. Rad, Lili Ostopovici, Cristian G. Bologa
UNM Division of Biocomputing
J. Comput. Aided Mol. Design 2007, 21:113–119
http://screening.health.unm.edu/supplements/LeadDrugPub2.csv
for supplementary materials
A Symposium to Honor Yvonne C. Martin
ACS Chicago, 25 March 2007
The University of New Mexico Health Sciences Center
Copyright © Tudor I. Oprea, 2007. All rights reserved
SCHOOL OF MEDICINE
Brief History (how I met Yvonne)
• First contact: EuroQSAR 1992 (Strasbourg), where she
expressed interest in one of my posters
• Continuing contact: EuroQSARs, Gordon Conferences
(QSAR/CADD), MUGs, CUPs
• Yvonne loves to visit New Mexico
• I followed her in leading the QSAR Society
• (in style, too)
• We renamed it The Cheminformatics and QSAR Society
• Disclosure: I have yet to visit Abbott Labs
The University of New Mexico
SCHOOL OF MEDICINE
The NIH Roadmap: Some Numbers
NIH Roadmap Initiative
Molecular Libraries Initiative
4 Chemical Synthesis
Centers
MLSCN (9+1)
9 external centers
1 NIH intramural
20 x 10 = 200 assays
PubChem
(NLM)
ECCR (6) Predictive
Exploratory ADMET
Centers
(8)
CombiChem
Parallel synthesis
DOS
4 centers + DPI
100k–500k compounds
SAR matrix
250-300 thousand
small molecules
OUTPUT:
Chemical
Probes
Hundreds of HTS Assays
Slide modified from Alex Tropsha (UNC)
The University of New Mexico
SCHOOL OF MEDICINE
So, what Is A Chemical Probe?
• Answer A: It has not been decided yet
• Answer B: A chemical probe is a somewhat selective,
somewhat potent (sub-micromolar?) structure that works on
the target / phenotypic assay of interest; it should be
reasonably …soluble (I guess)
• Chemical Probes are not necessarily anticipated to work in
vivo (though it will be preferred) and not expected to lead to
drugs either
• Chemical Probes could be used for assay development &
optimization, for imaging, radio-labeling, for flow cytometric
analyses, etc. (not necessarily used to query biological
space only)
The University of New Mexico
SCHOOL OF MEDICINE
Thus,
Why Bother with Drug Properties?
• Drug Discovery scientists sometimes think that
the name of the game is to get high affinity to
the target receptor, and then…
game over
• Not quite…
The University of New Mexico
SCHOOL OF MEDICINE
Neuraminidase Inhibitors for Influenza
•
X-ray structure guided rational design
•
•
GRID-suggested replacing -OH with basic functionality
Physical properties not amenable for oral delivery
Nature 1993, 363, 418.
NH2
OH
NH
HN
O
HO
N
H
CO2H
O
N
H
O
CO2H
O
HO
HO
OH
IC50 = 8600nM
Lead molecule
•
•
HO
OH
IC50 = 5nM
Zanamivir
GSK markets this as Relenza,
First drug for influenza
Slide modified from Andy Davis (AstraZeneca R&D Charnwood)
Gilead Neuraminidase Inhibitors
NH2
O
N
H
CO2H
NH2
HO
O H
O
H
O
O
H
N
H
CO2H
O
O
Glu 276
IC50 = 150nM
IC50 = 1nM
• Zwitterion not amenable for oral delivery
• Ethyl ester (oseltamivir) good oral absorption, duration
• Marketed as Tamiflu, first oral drug for influenza
J. Am. Chem. Soc. 681, 119, 1997
J. Med. Chem. 2451, 41, 1998
Slide modified from Andy Davis (AstraZeneca R&D Charnwood)
Inhalation to Overcome Low Bioavailability
Relenza vs Tamiflu
• Both potent neuraminidase inhibitors
• Relenza: Zanamivir delivered by using Diskhaler
• Tamiflu simple tablet formulation
•
Deesterified in plasma long plasma T½
• Tamiflu (marketed by Roche)
•
took 65% U.S. market-share from Relenza in 7 weeks
• Q1/Q2 2002 sales Relenza vs Tamiflu
•
Relenza market share fallen to 10%
•
GSK quoted reason “Slowness of the US to adopt
inhalation therapies”
Slide modified from Andy Davis (AstraZeneca R&D Charnwood)
Holistic Drug Design
Blood potency
1 dose a day ?
Fold over pA2
2 doses a day ?
MEC=( Ki x ff x 3)
Bioavailability
VDss and Clearance
24
MEC.VDss exp( kel ) 1
Predicted Human Dose (mg/kg/day )
%Oral
< 3mg/kg/day
200 mg
300mg/kg/day
20000 mg
Slide modified from Andy Davis (AstraZeneca R&D Charnwood)
MedChem Space & CLogP
A. Hopkins et al: LE = ΔG/N_at > 0.3
Free Energy of Binding (≈1.42*-log(Act)
20
18
16
14
12
99,119 out of 135,673
records have LE ≥ 0.3.
10
Of these, 78,393 have
activity ≥ 1 μM, and
29,980 activity ≥ 10 nM.
8
cLogP < 0
0 < cLogP < 4.5
cLogP > 4.5
6
4
1,310
17,389
11,281
2
10
20
30
Nr. heavy atoms (N_at)
40
50
The University of New Mexico
SCHOOL OF MEDICINE
The Mis-Use of RO5 Scores
• Pharmaceutical lead
80%
ACD
70%
MDDR
PDR
discovery world-wide apply
Lipinski’s Rule of 5: MW ≤
500, cLogP ≤ 5, HDO ≤ 5,
HAC ≤ 10. Any two
violations = poor %Oral
• Ro5 does not discriminate
20%
10%
0%
PASS
FAIL
SKIPPED
T.I. Oprea, J Comput-Aided Mol Des 2000 14, 251-264
“druglikeness”. Its use is
intended as filter in early
HTS hit analysis/discovery.
Problem is, it is applied
literally (but was derived
from drugs, not leads).
The University of New Mexico
SCHOOL OF MEDICINE
•
Common Sources for Leads(*) in
Drug Discovery
(*) Chemical Probes in the NIH Roadmap
Easier to optimise
Leadlike leads
affinity > 0.1 mM
MW ≤ 300
CLogP ≤ 3.0
High-affinity leads
affinity << 0.1 mM
MW >> 450
CLogP < 4.5
DRUG
Serendipity
Difficult to optimise
Druglike leads
affinity > 0.1 mM
MW > 450
CLogP > 4.5
One needs to distinguish “leadlike” leads from other sources of lead
structures, e.g., natural products that are high-affinity compounds (NPY
or taxol are leads!) or from “druglike” leads that are marketed
structures (e.g., salbutamol or HTS actives from “normal” combichem)
S. Teague et al., Angew. Chem., 1999, 38, 3743-3748
The University of New Mexico
SCHOOL OF MEDICINE
4 Generations of Progesterone Derivatives
O
O
O
O
O
H
H
O
H
H
O
H
H
H
H
O
O
Cl
H
H
H
O
O
Cyproterone
acetate
(1974)
Medroxyprogesterone acetate
(1958)
H
H
H
H
O
Progesterone
(1934)
O
H
O
H
H
H
O
H
H
H
Etonogestrel
H
H
H
O
O
Norethindrone
(1958)
Levonorgestrel
(1978)
H
H
Desogestrel
(1982)
The University of New Mexico
SCHOOL OF MEDICINE
The First Synthetic Drugs…
N
N
O
O
O
N
O
N
O
O
O
N
O
O
O
O
Cocaine
(1884)
Cl
N
N
O
N
O
Procainamide
(1953)
N
Coniine
(Socrates)
O
N
F
Cl
Cisapride
(1986)
O
O
O
N
O
O
Amylocaine
(1902)
Orthocaine
(1896)
N
N
N
O
Metoclopramide
(1964)
O
O
Br
N
N
N
O
O
Nirvanin
(1898)
Procaine
(1910)
O
N
N
Remoxipiride
(1988?)
The University of New Mexico
SCHOOL OF MEDICINE
More Synthetic Drugs…
S
O
N
N
O
O
O
N
Cl
N
N
N
O
O
O
N
O
Gravitol
(1929)
Orthocaine
(1896)
O
O
O
N
O
N
N
N
Cl
O
O
N
Diazepam
(1963)
N
O
O
Cocaine
(1884)
Chlorpromazine
(1952)
O
Cl
Prosympal
(1933)
Oxazepam
(1965?)
+
N
Cl
O
O
O
S
S
N
N
Nirvanin
(1898)
Prometazine
(1946)
N
Chlordiazepoxide
(1960)
N
O
O
O
N
Diltiazem
(1981?)
The University of New Mexico
SCHOOL OF MEDICINE
Leads 4: H2 Blockers
N
N
N
N
Interferes with drug
(P450) metabolism
N
N-alpha-guanyl histamine
Consequences…
1.
2.
N
N
Tautomerism
believedas
to nr.1
be besttoxic
Tagamet® is replaced
by Zantac®
essential
S
selling drug
pKa < 3
N
Income
nr.1 in topN 10 N
N Zantac® boosts Glaxo to S
N from
pharma
N
N
Burimamide
3.
N
Cimetidine
Glaxo acquires SmithKline Beecham
Isosteric
… all because of an imidazole-excessive
replacementpatent coverage
N
S
+
N
S
O
N
N
AH1866
pKa < 3
O
S
N
O
N
T. I. Oprea et al., J. Chem. Inf. Comput. Sci., 2001, 41, 1308-1315
N
O
Ranitidine
The University of New Mexico
SCHOOL OF MEDICINE
People always think that “newer drugs”
have higher MW than “older drugs”
400
300
200
100
1990
1980
1970
1960
1950
1940
0
1930
MEAN MOLECULAR
WEIGHT
Mean molecular weight against year of
introduction for oral marketed drugs
DECADE
• MW increase: ~50dalton/40 years
• The Property Space applies to most of us
Slide from Andy Davis, AstraZeneca Charnwood
The University of New Mexico
SCHOOL OF MEDICINE
Current Chemical Space Sampling
500000
400000
Well-sampled
300000 chemical space
Under-sampled
chemical space
Compounds with
given MW
Cumulative
(observed)
200000
Cumulative
(exponential
estimate)
100000
0
0
200
400
600
800
MW (a.m.u.)
M.M. Hann & T.I. Oprea, Curr. Opin. Chem. Biol., 2004, 8, 255-263
The University of New Mexico
SCHOOL OF MEDICINE
Is There a Preferred Property Space
for Leads & Chemical Probes?
• There is a tendency in the academic sector to
ignore past mistakes from the pharmaceutical
industry, e.g., the “tyranny of Lipinski” and “we
don’t care about in vivo, we just want chemical
probes”, which is unfortunate…
since the output of academic research ought to
result in tools to better understand biology
(pharmacology, chemical biology, etc)
• So the reason for this talk is to learn from drug
discovery, and from the failures in the pharma
sector?
T. I. Oprea et al., J. Chem. Inf. Comput. Sci., 2001, 41, 1308-1315; updated
The University of New Mexico
SCHOOL OF MEDICINE
Leads and Actives Datasets
• 385 leads and the 541 drugs that emerged from these
leads, which resulted by combining previously described
datasets (Hann et al., Proudfoot, Oprea et al)
• Compounds of current interest extracted from PubChem,
categorized according to their source and PubChem activity
label, as follows:
•
152 ‘‘actives’’ from MLSMR and MLSCN, referred to as MLSMR
Act;
•
46 ‘‘actives’’ from Nature Chemical Biology, tested in MLSCN,
referred to as NCB Act;
•
1,488 ‘‘inactives’’ from MLSMR and MLSCN, referred to as
MLSMR Inact;
•
72 ‘‘inactives’’ from Nature Chemical Biology, tested in MLSCN,
referred to as NCB Inact;
T. I. Oprea et al. J. Comput. Aided Mol. Design 21:113-119, 2007
The University of New Mexico
SCHOOL OF MEDICINE
Drugs & Bio-Activity Datasets
• Compounds in pharmaceutical development, extracted from
the MDDR (MDL Drug Data Report) 2005.2 database,
categorized according to their clinical testing phase, in the
following manner:
•
•
•
•
1,147 launched drugs;
301 compounds in phase III clinical trials, referred to as Phase III;
1,047 compounds in phase II clinical trials referred to as Phase II;
801 compounds in phase I clinical trials, referred to as Phase I;
• Compounds extracted from WOMBAT 2006.1, which indexes
papers published in mainstream medicinal chemistry
journals, split in 2 categories:
•
•
30,690 compounds for which the biological activity is above 1 lM, or
below 6 units on the –log10 (activity) scale, on all of the documented
literature assays (WB6);
5,784 compounds for which the biological activity is below 1 nM, or
above 9 units on the –log10 (activity) scale, in one of the
documented literature assays (WB9). Of these, only 127 were
launched drugs.
T. I. Oprea et al. J. Comput. Aided Mol. Design 21:113-119, 2007
The University of New Mexico
SCHOOL OF MEDICINE
WOMBAT 2006.1
•
WOMBAT 2006.1 contains 154,236 entries (136,091 unique SMILES),
totaling 307,700 biological activities on over 1,320 unique targets.
•
WOMBAT 2006.1 contains 6,801 different series from 6,791 papers
published in medicinal chemistry journals between 1975 and 2005
•
Systematic coverage for: J. Med. Chem. (77.6%) 1991-2004 [complete],
2005 [partial], Bioorg. Med. Chem. Lett. (15.4%) 2002-2003 [complete],
2004 [partial], Bioorg. Med. Chem. (5.6%) 2002-2003 [complete], Eur. J.
Med. Chem. (1%) 2002-2003 [complete] 2004 [partial];
•
SwissProt IDs for ~88% of the Entries
•
DOI (digital object identifier) links & PubMed IDs for all references (direct
access to PDF files for institutions with appropriate subscriptions)
•
ClogP and XMR from Biobyte Corporation (Al Leo), AlogP and LogSw from
ALOGPS (Igor Tetko), Ligand Efficiency, Rule-of-Five, and Molecular
Complexity can be queried.
•
Over 10,000 unique entries are added every six months
M. Olah et al. Chemical Biology, Wiley-VCH 2007, in press
Databases Scanned for Molecular Properties
Type
Leads
SMR MLSCN Act
NCB Act
WB9
Drugs
MDDR_Launched
MDDR_Phase I
MDDR_Phase II
MDDR_Phase III
SMR MLSCN Inact
NCB Inact
WB6
Count
385
152
46
5,784
541
1,147
808
1,061
303
1,488
72
30,690
Source; Comments
From Hann, Proudfoot, Oprea datasets
MLSMR; Actives only in MLSCN assays
Nature Chem Biol; Actives in any assay
WOMBAT 2006.1; nM compounds
Marketed drugs from the above Leads set
MDDR 2005.2, all launched drugs
MDDR 2005.2, all phase I candidate drugs
MDDR 2005.2, all phase II candidate drugs
MDDR 2005.2, all phase III candidate drugs
MLSMR; Inactives only in MLSCN assays
Nature Chem Biol; Inactives in any assay
WOMBAT 2006.1; mM compounds
Note: PubChem queries were performed in August 2006; some of the
structures & “active” definitions may have changed – this is an
evolving database, and users sometimes revise published data
T. I. Oprea et al. J. Comput. Aided Mol. Design 21:113-119, 2007
The University of New Mexico
SCHOOL OF MEDICINE
7 11 3 15 2
1 19 21 4
685
Interpretation
Leads, Drugs, MDDR, WOMBAT midpoints
Type
Leads
SMR MLSCN Act
NCB Act
WB9
Drugs
MDDR_Launched
MDDR_Phase I
MDDR_Phase II
MDDR_Phase III
SMR MLSCN Inact
NCB Inact
WB 6Only
Count
385
152
46
5,784
541
1,147
808
1,061
303
1,488
72
30,690
MW SMCM RNG HAC RTB CLP TLogP TLogSw Ro5
287.4
37.6
3Nr of
4 flex.
4 2.3
2.3
-3.3
0
Complexity
273.9
30.0
3bonds
4
4 2.5
2.5
-3.3
0
(R2=0.55
273.9 37.2
3
3
2 3.1
3.2
-3.8
0
2 methods
against
MW)
463.6 56.7
4
6 10 3.8
3.6
-4.7
1
LogP2.6
source
code
oct
332.7
3
4
6for2.6
-3.8
0
Nr43.3
of RINGS
available
345.4
42.7
3
5
6 2.3
2.4
-3.7
0
LogS
Nr of H-bond
wat:
420.6
51.3
3
6
8 3.0
2.9
-4.3
0
from UNM
“intrinsic”
acceptors;
399.5 49.2
3
6no 8 3.2
2.9
-4.3
0
aqueous
significant
378.9 46.4
3
5change
7 2.7
2.6
-4.0
0
260.3 28.5
2
4
4 2.0
2.0
-3.0
0
solubility
in H-bond
donors
254.8 31.2
2
4
3 1.6
1.7
-2.6
0
364.4 42.6
3
5
6 3.0
2.9
-4.2
0
All values are the median (50% distribution moment)
T. I. Oprea et al. J. Comput. Aided Mol. Design 21:113-119, 2007
The University of New Mexico
SCHOOL OF MEDICINE
Actives, Drugs, WB9 & Inactives –
Median & Tail Values
Type
Actives 50
Actives 90
Drugs 50
Drugs 90
WB 9 Mul 50
WB 9 M ul 90
Inactives 50
Inactives 90
Count
569
569
1,651
1,651
5,784
5,784
32,114
32,114
MW SMCM RNG HAC RTB CLogP TLogPTLogSw Ro5
284.3 35.2
3
4
4
2.5
2.4
-3.3
0
432.5 59.8
4
8
9
5.1
4.6
-5.1
1
339.5 42.6
3
5
6
2.3
2.3
-3.7
0
558.9 73.2
5 12 14
5.3
4.8
-5.5
2
463.6 56.7
4
6 10
3.8
3.6
-4.7
1
761.1 94.7
6 14 25
6.5
5.6
-6.0
2
358.4 41.8
3
5
6
2.9
2.8
-4.1
0
581.7 70.0
5 11 17
6.0
5.2
-5.8
2
•
All “90” values are the 90% distribution moment, except TLogSw (@ 10%)
•
Actives are less complex, less flexible, slightly more soluble than drugs
•
WB9 (literature) actives are more complex, more flexible, more
hydrophobic & less soluble when compared to actives & drugs
T. I. Oprea et al. J. Comput. Aided Mol. Design 21:113-119, 2007
The University of New Mexico
SCHOOL OF MEDICINE
Actives, Drugs, WB9 & Inactives –
Change from Median & Tail Values
Type
Actives 50
Actives 90
Drugs 50
Drugs 90
WB 9 Mul 50
WB 9 Mul 90
Inactives 50
Inactives 90
Count
569
569
1,651
1,615
5,784
5,784
32,114
32,114
MW SMCM RNG HAC RTB CLogP TLogPTLogSw Ro5
284.3 35.2
3
4
4
2.5
2.4
-3.3
0
432.5 59.8
4
8
9
5.1
4.6
-5.1
1
55.2
7.4
0
1
2
-0.2
-0.1
-0.4
0
126.4 13.4
1
4
5
0.3
0.2
-0.3
1
179.3 21.5
1
2
6
1.4
1.2
-1.4
1
328.6 34.9
2
6 16
1.4
1.0
-0.9
1
74.1
6.6
0
1
2
0.4
0.4
-0.8
0
149.2 10.2
1
3
8
1.0
0.6
-0.7
1
•
This analysis can give us trends & boundaries for this property space,
e.g., we could use tail-end values from the above to filter for lead- or
“pub”-like-ness (is there a preferred space for chemical probes?)
•
Learn from the WOMBAT “nM” ligands: these are failed medchem
projects from big pharma (i.e., not too good as leads). Data very useful
for ideas, but I would not advise anyone to use these as leads
•
Note: if working with natural products, these filters will fail
T. I. Oprea et al. J. Comput. Aided Mol. Design 21:113-119, 2007
The University of New Mexico
SCHOOL OF MEDICINE
Understanding the Property Space for Actives
• 100 MW 432.5, -4 ClogP 5.1, LogSw ≥ -5.1, *Hdon 4,
*Hacc 9, *SMCM 60, *RTB 9, *RNG 4 (*low value 0):
• This filter covers 68.5% of the actives, 50.5% of the drugs,
17.7% of the WB nM ligands, and 49.8% of the WB μM ligands.
• 100 MW 432.5, -4 ClogP 5.1, LogSw ≥ -5.1, *Hdon 4,
*Hacc 9, *SMCM 73.2, *RTB 14, *RNG 5 (low value 0):
• This filter covers 75% of the actives, 59.7% of the drugs, 26.4%
of the WB nM ligands, and 54.5% of the WB μM ligands.
• 100 MW 559, -4 ClogP 5.3, LogSw ≥ -5.5, *Hdon 5,
*Hacc 12, *SMCM 73.2, *RTB 14, *RNG 5 (low value 0):
• This filter covers 81.4% of the actives, 69.3% of the drugs,
46.6% of the WB nM ligands, and 64.8% of the WB μM ligands.
•
At ClogP > 4.1 & LogSw < -4.1, we find 15.1% of the actives, 19.4% of the
drugs, 39.5% of the WB nM ligands, and 27.8% of the WB μM ligands
•
Obs: none of the above combinations yields Ro5 ≥ 2 violations!!!
The University of New Mexico
SCHOOL OF MEDICINE
(Revised) Guidelines for Probe Discovery
•
The following (restrictive!) properties could be considered when evaluating
chemical probes, in particular when many HTS primary hits come out:
• 100 MW 432.5, -4 ClogP 5.1, LogSw ≥ -5.1,
0 Hdon 4, 0 Hacc 9 (inspired from the tail-end of Actives)
• Obs.: MW cut-off in Ro5 is 427, according to Michal Vieth
• SMCM 73.2, RTB 14, RNG 5 (from the tail-end of Drugs)
•
Good probes require that subtle interplay between solubility and
permeability, in order to work in cells and in vivo.
• For further progression (lead opt.), additional properties are required:
• %F ≥ 30, CL 30 mL/min, %PPB 99 (in rat PK models)
• KD ≥ 100 mM for drug-metabolizing P450s (no DDIs)
• Preferably, no acute toxicity, no carcinogenicity, etc.
•
These cut-off values will change with each target, its location (e.g., brain vs.
stomach vs. bone vs. kidney), with the intended admin. mode…
These values are not the pharma equivalent of the Planck constant
The University of New Mexico
SCHOOL OF MEDICINE
Conclusions
• The MLSMR & NCB actives (N=198) has similar property
distribution with the historical Leads (N=385).
• It’s interesting to compare the differences between the
property distribution values of the 569 Actives and the
5,784 high-activity molecules (WB9).
• The WB9 subset contains molecules that are, on average,
larger, more hydrophobic and less soluble than any of the
other datasets examined here.
• Pub-like Actives are, on average, smaller, less complex,
less hydrophobic and more soluble than the other
datasets.
The University of New Mexico
SCHOOL OF MEDICINE
Conclusions 2
• As we discover new chemical probes, the issue of what
constitutes high-quality probes is under scrutiny.
• Arguments such as ‘‘historical bias’’ are used when it comes
to defining property boundaries (the ‘tyranny of Lipinski’).
• Yet, the number of new approved drugs /year continued to
decline in the past decade, despite significantly larger
numbers of molecules & targets explored.
• Whether the boundary limits will be extend beyond the Ro5
‘‘cube’’, only time will tell.
• Over 55% of the top 200 oral drug products in the United
States, Great Britain, Japan and Spain are ‘‘high-solubility
drugs’’ [*], and that only 18 of the 133 active principles from
these drugs have ClogP values greater than 4.0.
[*] Takagi T et al., Mol. Pharmaceutics 2007, 3:631
The University of New Mexico
SCHOOL OF MEDICINE
In Fairness: Computer Aided Drug Design
Marketed drugs whose discovery was aided by computers
Generic
name
Brand
Name
US
Approval
CADD
Method
Therapeutic
Category
Norfloxacin
Noroxin
1983
QSAR
Antibacterial
Losartan
Cozaar
1994
CADD
Antihypertensive
Dorzolamide
Trusopt
1995
CADD
Antiglaucoma
Ritonavir
Norvir
1996
CADD
Antiviral
Indinavir
Crixivan
1997
QSAR
Antiviral
Donepezil
Aricept
1997
QSAR
Anti-Alzheimer's
Zolmitriptan
Zomig
1997
CADD
Antimigraine
Nelfinavir
Viracept
1997
SBDD
Antiviral
Amprenavir
Agenerase
1999
SBDD
Antiviral
Zanamivir
Relenza
1999
SBDD
Antiviral
Oseltamivir
Tamiflu
1999
SBDD
Antiviral
Lopinavir
Aluviran
2000
SBDD
Antiviral
Imatinib
Gleevec
2001
SBDD
Antineoplastic
Erlotinib
Tarceva
2004
SBDD
Antineoplastic
Ximelagatran
Exanta
2004 (EU)
SBDD
Anticoagulant
The University of New Mexico
SCHOOL OF MEDICINE
Acknowledgments
• Simon Teague, Andy Davis and Paul Leeson (AstraZeneca
R&D Charnwood), Mike Hann, Andrew Leach and Gavin
Harper (GSK Medicines Research Centre, Stevenage), and
John Proudfoot (Boehringer Ingelheim, Ridgefield CT)
worked on the leadlike concept
• WOMBAT Team: Maria Mracec, Marius Olah, Lili
Ostopovici, Ramona Rad, Alina Bora, Nicoleta Hadaruga,
Ramona Moldovan, Dan Hadaruga (Romanian Academy
Institute of Chemistry, Timisoara, Romania)
• Funding:
• NIH U54 MH074425-01
The University of New Mexico
SCHOOL OF MEDICINE
Yvonne Likes To Keep Her Eyes Open
• Here, giving her “grandma” talk
The University of New Mexico
SCHOOL OF MEDICINE
QSAR Reborn
A Symposium in Honor of Philip Magee
• Dr. Phil S. Magee (1926-2005) was one of the
•
•
•
•
pioneers in utilizing QSAR, mostly in agrochemistry
and transdermal property modeling. Served as first
Chair of the QSAR and Modeling Society
Symposium organized by Bob Clark, John Block,
Lowell Hall & Lermont Kier
At ACS Boston, August 2007, sponsored by the
COMP, CINF and AGRO Divisions
Deadline for Abstracts: April 2, 2007
Topics: QSAR Descriptors, QSAR Techniques, and
QSAR Applications
The University of New Mexico
SCHOOL OF MEDICINE